The U.S. Geological Survey Ohio Water Microbiology Laboratory
Links
- Document: Report (4.28 MB pdf) , HTML , XML
- Download citation as: RIS | Dublin Core
The mission of the Ohio Water Microbiology Laboratory is to provide microbiological data of public health significance from surface waters, groundwaters, and sediments for a variety of study objectives.
Get To Know the Ohio Water Microbiology Laboratory
The U.S. Geological Survey Ohio Water Microbiology Laboratory (OWML) is a part of the Ohio-Kentucky-Indiana Water Science Center. The laboratory conducts internal projects, works with external cooperators, and assists U.S. Geological Survey offices and National programs. The laboratory offers guidance, study design, and data interpretation expertise to collaborators, all following rigorous quality control and quality assurance procedures.

Photograph of U.S. Geological Survey biologists and technicians working in the Ohio Water Microbiology Laboratory. Photograph by Amie Brady, U.S. Geological Survey.
Laboratory Capabilities
The Ohio Water Microbiology Laboratory is a Biosafety Level II water research facility located in Columbus, Ohio. The laboratory scientists specialize in the detection of microbiological indicators and pathogens and also study the processes that affect the occurrence and distribution of these constituents in the environment. The laboratory works with other scientists in the U.S. Geological Survey as well as external cooperators including other Federal, State, and local government agencies, academic institutions, and public utilities. The laboratory provides assistance for projects related to various topics, including harmful algal blooms, predictive modeling for recreational water quality and source-water quality, rapid microbial detection methods, and microbial source tracking. The laboratory offers a wide range of analytical services and maintains rigorous quality assurance practices for all processes to ensure traceability and reliability of all results. All services are compliant with the U.S. Geological Survey Quality Management System, and samples are tracked from arrival in the laboratory to dissemination of results to cooperators.
Indicators of Fecal Contamination
Fecal indicator organisms (both bacteria and viruses) are used to determine the potential contamination of water by fecal material. Detection of fecal indicator organisms suggests that harmful organisms may be present. Culture-based (fig. 1) and molecular-based methods can be used to determine concentrations of these organisms to understand the risk associated with using these waters for recreation or as drinking water sources. The Ohio Water Microbiology Laboratory has the capability to analyze for a wide range of fecal indicator organisms, which are described in the following list.
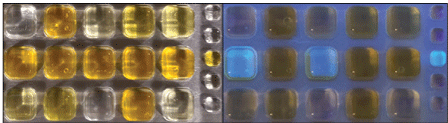
Photographs of detected total coliforms (yellow under ambient light, left) and Escherichia coli (fluorescent under ultraviolet light, right) using a Colilert method from IDEXX Laboratories. Photograph by Braden Lanier, U.S. Geological Survey.
-
• Coliform bacteria are a group of bacteria found in soil, water, vegetation, and the intestines of humans and animals. Concentration of total coliforms in water can be used as a general indicator of bacterial contamination.
-
• Escherichia coli is a species of coliform bacteria and a natural resident of the intestines of warm-blooded animals. Concentrations of E. coli can be used as direct evidence of fecal contamination.
-
• Enterococcus is a genus of bacteria commonly found in the intestines of warm-blooded animals. Enterococcus bacteria are more persistent in water than coliforms. Enterococcus concentrations can be used as a measure of recreational water quality and can be used to track the movement of fecal contamination in groundwater.
-
• A coliphage is a type of virus that require coliform bacteria to replicate. Coliphages may be used as fecal indicator organisms but may more accurately show the survival and transport of viruses than bacteria. These viruses are detected using viral culture methods.
Microbial Source Tracking
Fecal contamination can come from a variety of potential sources, and traditional measures of assessment, such as culture-based methods, do not provide source-specific information. Microbial source tracking (MST) techniques can be used to improve the understanding of contamination sources (fig. 2). For example, warm-blooded animals have unique factors such as diet, temperature, and physiology that select for specific gut microbial populations in their intestines. These host-specific microbes contain unique genetic sequences that are shed into the environment. In a microbial source tracking workflow (fig. 2), water bodies of interest are first sampled and filtered to catch any microbes in the water. The deoxyribonucleic acid (DNA), a type of genetic material, is then extracted from microbes caught on the filter. This DNA is then put through a specific series of quantitative polymerase chain reaction (qPCR) assays, which allows the laboratory to determine which fecal sources were present in the sample. qPCR methods are used to detect and quantify host-specific genetic sequences or microbial source tracking markers for fecal contamination from humans, dogs, cattle, horses, pigs, or birds.
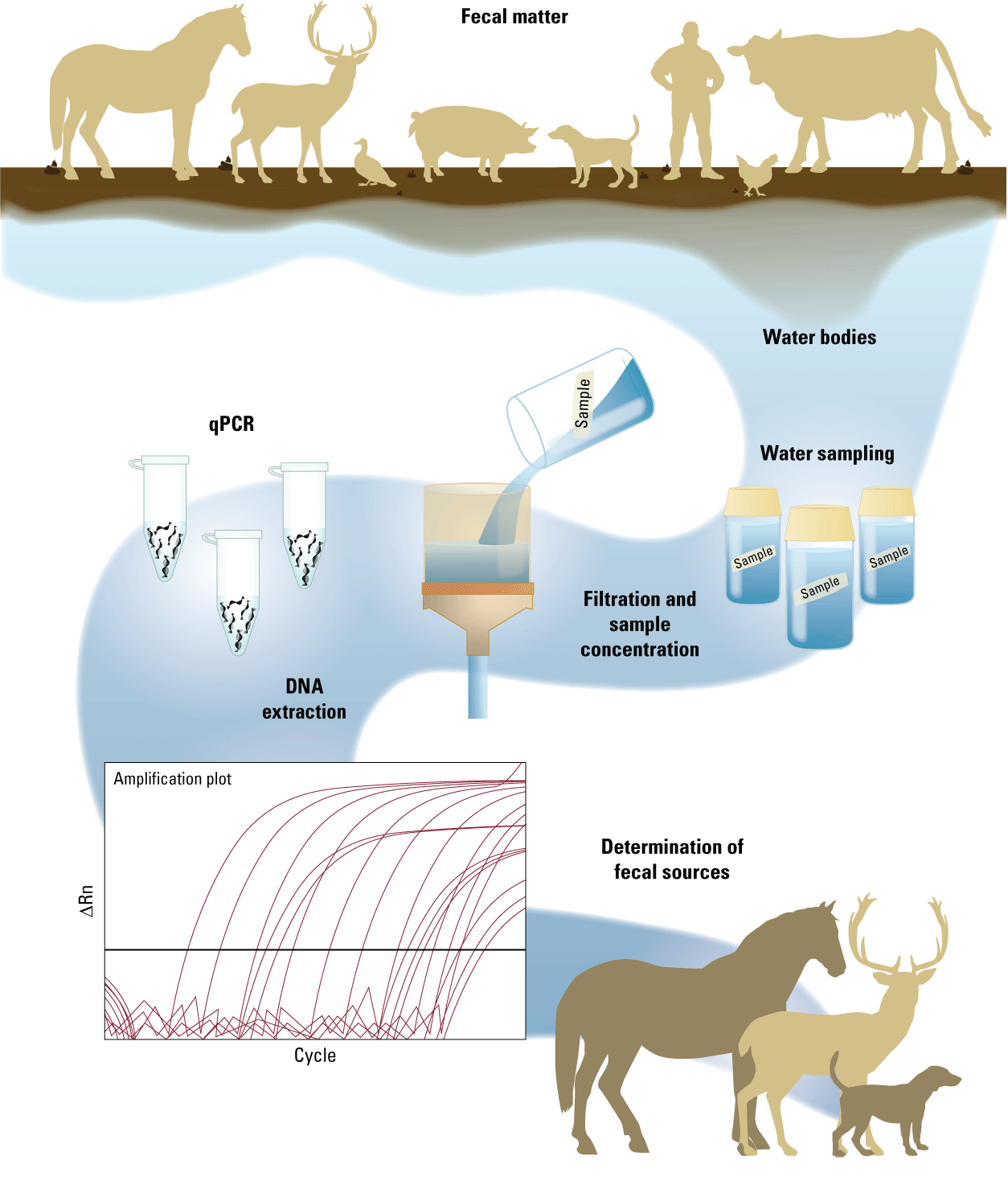
Diagram of a microbial source tracking (MST) workflow. DNA, deoxyribonucleic acid; qPCR, quantitative polymerase chain reaction; ΔRn, normalized reporter value.
Environmental Deoxyribonucleic Acid (eDNA) Detection
Environmental deoxyribonucleic acid (eDNA) is genetic material shed into the environment from a live organism. The eDNA can be used to identify the presence of the organism without capturing the organism in question. Sloughed tissue (such as skin or mucus cells), gametes, excrement, and shell material can all be used as eDNA targets. eDNA studies can help with rare species detection, invasive species tracking, and can also be used to assess the health of streams and wetlands. The Ohio Water Microbiology Laboratory uses qPCR methods to detect eDNA targets for a variety of aquatic animals, including several species of invasive carp, several threatened or endangered mussels, and some macroinvertebrates.
Cyanobacterial Harmful Algal Blooms
The increasing prevalence of cyanobacterial harmful algal blooms (also known as cyanoHABs) and the toxins they produce are a global water-quality issue that threatens human and wildlife health (fig. 3). Additional monitoring of toxins in drinking source and recreational waters may be required if a cyanobacterial harmful algal bloom is present.

Photograph of a cyanobacterial harmful algal bloom at Maumee Bay State Park, City of Oregon, Ohio, in 2013. Photograph by Donna Francy, U.S. Geological Survey.
Even though the name “algal blooms” refers to algae, cyanobacterial harmful algal blooms do not involve organisms that are traditionally considered algae. Cyanobacteria are a group of bacteria that are commonly found in freshwater and can produce their own energy through photosynthesis. Importantly, some species of cyanobacteria can produce toxins harmful to humans and animals, known as cyanotoxins. Cyanotoxins can be quite potent and include neurotoxins, hepatotoxins, cytotoxins, and endotoxins.
At the laboratory, cyanobacterial genes are detected by qPCR assays. Knowing the specific genes can help identify what types of cyanobacteria are present and whether they have the potential to produce toxins. Enzyme-linked immunosorbent assays (ELISAs) are also used by the laboratory to directly identify and analyze cyanobacteria toxins.
Taste and Odor Issues in Water
Cyanobacteria and filamentous bacteria in the genus Actinomyces can produce odorous chemicals that can affect drinking water quality. These chemicals are not harmful but are considered nuisance constituents in water and can cause some consumers to think that their drinking water is unsafe. Both culture-based and qPCR methods can be used to detect genes that produce nuisance compounds.

Photograph of a girl drinking water with an earthy taste and smell. Photograph by Erin Stelzer, U.S. Geological Survey.
Cyanobacteria and Actinomyces are typically found in soil and water and can produce the compounds geosmin and 2-methylisoborneol (MIB), which are responsible for earthy taste and odor in water.
Enumeration of Actinomyces is done by culture and standard plate counts. Additionally, qPCR assays are used to detect genes that synthesize these earthy compounds in both cyanobacteria and Actinomyces bacteria.
Predictive Modeling
Risks associated with recreation in natural waters include illness caused by pathogens from fecal contamination and cyanotoxins produced by cyanobacteria. Swim advisories or beach closings issued by recreational water managers are based on the concentrations of fecal indicator organisms such as Enterococcus and E. coli, or cyanotoxin concentrations. Traditional culture-based methods for determining fecal indicator bacterial concentrations are slow and take at least 18 to 24 hours for results to be available (fig. 1). Cyanotoxin concentrations can be determined within hours but require a trained technician and specialized laboratory equipment. This means that sample results may take too long for any meaningful warning or beach closure that would protect public health. Predictive modeling solves this problem by using past data to estimate water-quality conditions in near-real time. Analysis by the laboratory can be used for real-time forecasting. Concentration of fecal indicator organisms determined by the laboratory are combined with real-time measured variables like temperature and precipitation to determine a predicted bacterial concentration. The predicted value is then used to decide if water quality is adequate for public recreation. Scientists at the Ohio Water Microbiology Laboratory have developed models for determining the concentration of cyanotoxin, fecal indicator organisms, and Enterococci. Fecal indicator predictive models can be created with a variety of environmental variables such as rainfall and turbidity.
For more information, visit the laboratory website at https://www.usgs.gov/labs/ohio-water-microbiology-laboratory, or contact us by email at gs-w-ohclb_owml@usgs.gov.
Disclaimers
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Although this information product, for the most part, is in the public domain, it also may contain copyrighted materials as noted in the text. Permission to reproduce copyrighted items must be secured from the copyright owner.
Suggested Citation
Lanier, B.M., Brady, A.M.G., Cicale, J.R., Kephart, C.M., Lynch, L.D., Schroeder, M.W., and Stelzer, E.A., 2024, The U.S. Geological Survey Ohio Water Microbiology Laboratory: U.S. Geological Survey Fact Sheet 2024–3004, 4 p., https://doi.org/10.3133/fs20243004.
ISSN: 2327-6932 (online)
| Publication type | Report |
|---|---|
| Publication Subtype | USGS Numbered Series |
| Title | The U.S. Geological Survey Ohio Water Microbiology Laboratory |
| Series title | Fact Sheet |
| Series number | 2024-3004 |
| DOI | 10.3133/fs20243004 |
| Publication Date | June 14, 2024 |
| Year Published | 2024 |
| Language | English |
| Publisher | U.S. Geological Survey |
| Publisher location | Reston, VA |
| Contributing office(s) | Ohio-Kentucky-Indiana Water Science Center |
| Description | 4 p. |
| Online Only (Y/N) | Y |
| Additional Online Files (Y/N) | N |


