Genetic Population Assignments of Atlantic Sturgeon Provided to National Marine Fisheries Service, 2022
Links
- Document: Report (3.22 MB pdf) , HTML , XML
- Data Release: USGS data release - Individual assignments and microsatellite genotypes for Atlantic sturgeon from 2021
- Download citation as: RIS | Dublin Core
Acknowledgments
We thank E. Hilton, D. Gregg, J. Kahn, J. Gartland, T. Moore, H. Corbet, C. Stence, and members of the National Oceanic and Atmospheric Administration’s Northeast Fisheries Science Center for contributing samples used in this analysis. We also thank the many individuals and agencies who contributed to the genetic baseline used for assignment testing. Funding for this research was provided by the National Marine Fisheries Service to the USGS Eastern Ecological Science Center. We also thank the two anonymous reviewers for providing Fundamental Science Practices reviews.
Abstract
Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) were once abundant and supported large-scale fisheries throughout much of the east coast of the United States. However, historic overharvest and habitat loss resulted in dramatic declines in abundance and eventual listing under the Endangered Species Act of the United States. As part of this listing, Atlantic sturgeon populations were divided into five distinct population segments (DPSs), with many management activities occurring at the level of the DPS. However, because subadult and adult Atlantic sturgeon can make large, coast-wide migrations and often mix extensively with individuals from other populations, individuals may be exposed to conservation threats away from their natal river or DPS, ultimately making it difficult to determine the appropriate spatial scale for management activities. To help address this uncertainty, the U.S. Geological Survey performed genetic assignment tests to determine the natal origin of 329 Atlantic sturgeon that were encountered as mortalities or taken during permitted activities in 2021. Overall, most individuals assigned to the Hudson River population, with additional major contributions from the James River Fall and Delaware River populations. However, a sizeable proportion of individuals were assigned to more distantly located populations in the southeastern United States. These results highlight the prevalence of long-distance movements in Atlantic sturgeon and underscore that populations may be vulnerable to threats far from their natal rivers.
Introduction
Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) is an anadromous fish species that is broadly distributed from Labrador, Canada, to Florida, United States. Historic records suggest that Atlantic sturgeon may have once spawned in at least 35 rivers along the east coast of North America; however, today only 25 rivers are suspected to still support naturally reproducing populations (Atlantic Sturgeon Status Review Team, 2007; Atlantic States Marine Fisheries Commission, 2017). There is limited information regarding abundance (Wirgin and others, 2015), but it is generally believed that most populations are greatly reduced from historical sizes (Kazyak and others, 2020).
The decline of Atlantic sturgeon populations can largely be traced to an unsustainable commercial fishery that mostly collapsed in the late 19th century, but which continued through much of the 20th century despite severely reduced landings (Dadswell, 2006; Hilton and others, 2016). Following decades of population declines, the Atlantic States Marine Fisheries Commission enacted a 40-year moratorium on Atlantic sturgeon harvest in waters of the United States in 1998, with the goal of protecting 20-year classes of spawning females (Atlantic States Marine Fisheries Commission, 1998).
In 2012, the Atlantic sturgeon was listed under the Endangered Species Act of the United States (National Oceanic and Atmospheric Administration, 2012a, b). As part of this listing, and partially based on available genetic information at the time, rivers in the United States were divided into five distinct population segments (DPSs). Four DPSs were classified as endangered including the South Atlantic, Carolina, Chesapeake Bay, and New York Bight DPSs. The fifth DPS, the Gulf of Maine, was classified as threatened due to limited perceived threats to population persistence (Atlantic Sturgeon Status Review Team, 2007). In Canada, Atlantic sturgeon are designated as threatened by the Committee on the Status of Endangered Wildlife in Canada (2011). Within the United States, Federal management of Atlantic sturgeon is legally predicated on the DPS framework, with each DPS managed separately to limit the impacts of anthropogenic activities (such as dredging, offshore wind development, and commercial fisheries) on recovery. The Atlantic States Marine Fisheries Commission also seeks to evaluate stock status across multiple spatial scales, including range-wide, DPS-specific, and population-specific (Atlantic States Marine Fisheries Commission, 2017).
The need to monitor and manage threats to the recovery of specific stocks presents significant challenges to Atlantic sturgeon conservation. After hatching, juvenile Atlantic sturgeon remain in their natal tributary for up to six years before migrating to the ocean (Hilton and others, 2016). Once in marine waters, subadult and adult Atlantic sturgeon frequently engage in extensive coast-wide movements where behavior is not well-understood. However, telemetry data indicate that individuals can move several hundreds of kilometers (km) within a few months, with some individuals moving up to 1,500 km (Erickson and others, 2011; Rulifson and others, 2020). During these periods of extensive marine movements, it is common for high-density, mixed-stock aggregations to form in foraging areas (Dunton and others, 2010; Dadswell and others, 2016), and for individuals to temporarily occupy tidal sections of non-natal rivers and estuaries (Wirgin and others, 2015; Fox and others, 2018). Accordingly, because Atlantic sturgeon use a complex mosaic of habitats to complete their life cycle, they can be vulnerable to anthropogenic threats at both local and coast-wide scales (Erickson and others, 2011; Wirgin and others, 2015), including bycatch mortality (Dunton and others, 2015), marine construction, dredging, vessel strikes, dams, and potential predation by invasive species (Atlantic States Marine Fisheries Commission, 2017).
In order to mitigate population-specific exposure to human activities, it is necessary to understand how individuals from each population are distributed across the various habitats they use (Dunton and others, 2015). One tool that can be used to help meet this objective is individual-based genetic assignment tests, which use genotypic data to identify the likely natal population for an individual of unknown origin. Results of these individual-based genetic assignment tests can be used to help characterize the impacts of permitted activities on specific Atlantic sturgeon populations or the five DPSs listed under the Endangered Species Act. As part of an interagency agreement with the National Marine Fisheries Service (NMFS) that began 2018, the U.S. Geological Survey (USGS) has been performing individual-based genetic assignment tests for Atlantic sturgeon samples that have been submitted to the Atlantic Coast Sturgeon Tissue Research Repository (https://www.fisheries.noaa.gov/national/endangered-species-conservation/atlantic-coast-sturgeon-tissue-research-repository). This report provides results for samples obtained in 2021 and comments about the natal origin of individuals analyzed.
Methods
Tissue Collection and Genotyping
Each year, scientific collectors submit samples to the Atlantic Coast Sturgeon Tissue Research Repository and NMFS identifies samples that are of greatest interest for individual assignment testing. In 2021, NMFS identified 345 Atlantic sturgeon samples that were reported mortalities and/or necessary for assignment under Section 7 of the Endangered Species Act.
Samples were genotyped at the U.S. Geological Survey Eastern Ecological Science Center (USGS-EESC) in Kearneysville, West Virginia. Whole genomic DNA was extracted from tissue samples using Gentra Puregene reagents (Qiagen, Germantown, MD, USA). All samples were screened for 12 Atlantic sturgeon microsatellite loci (LS19, LS39, LS54, LS68, Aox12, Aox23, Aox45, AoxD170, AoxD188, AoxD165, AoxD44, AoxD241; described in May and others, 1997; King and others, 2001; Henderson-Arzapalo and King, 2002).
Sex was identified in 272 individuals by adding the AllWSex2 primer (Kuhl and others, 2021) into an existing PCR multiplex. Previous study (Nick Sard, SUNY Oswego, written commun., 2023) has shown that the amplicon base pair length and relative florescence units (RFU) can be used to identify individual sex, with results closely matching phenotypic and histological sex assignments. Specifically, amplicon fragments in females are approximately 106 base pairs and amplify with high intensity (greater than 26,000 RFU) whereas males produce no product or sometimes a faint band that is less than 8,000 RFU. These criteria were applied to determine the genetic sex of individuals. Occasionally the amplicon was greater than 8,000 RFU but less than 26,000, in which the sex was identified as unknown. If no loci amplified in the multiplex PCR with the sex marker, then the individual was categorized as unknown sex. All genotypes are available in a U.S. Geological Survey data release available at White and others (2023).
Data Analysis
Individual-based assignments were performed by assigning individuals to one of 18 genetically distinct populations represented in the genetic baseline for Atlantic sturgeon described by White and others (2021). The genetic baseline included 2,510 individuals collected from 13 rivers and one estuary. Based on significant genetic differentiation and previous genetic and behavioral studies, Atlantic sturgeon from the James, Pee Dee, Edisto, and Ogeechee rivers were divided into spring and fall runs for a total of 18 potential source populations. These populations were distributed across all five DPSs and included two Canadian rivers (fig. 1).
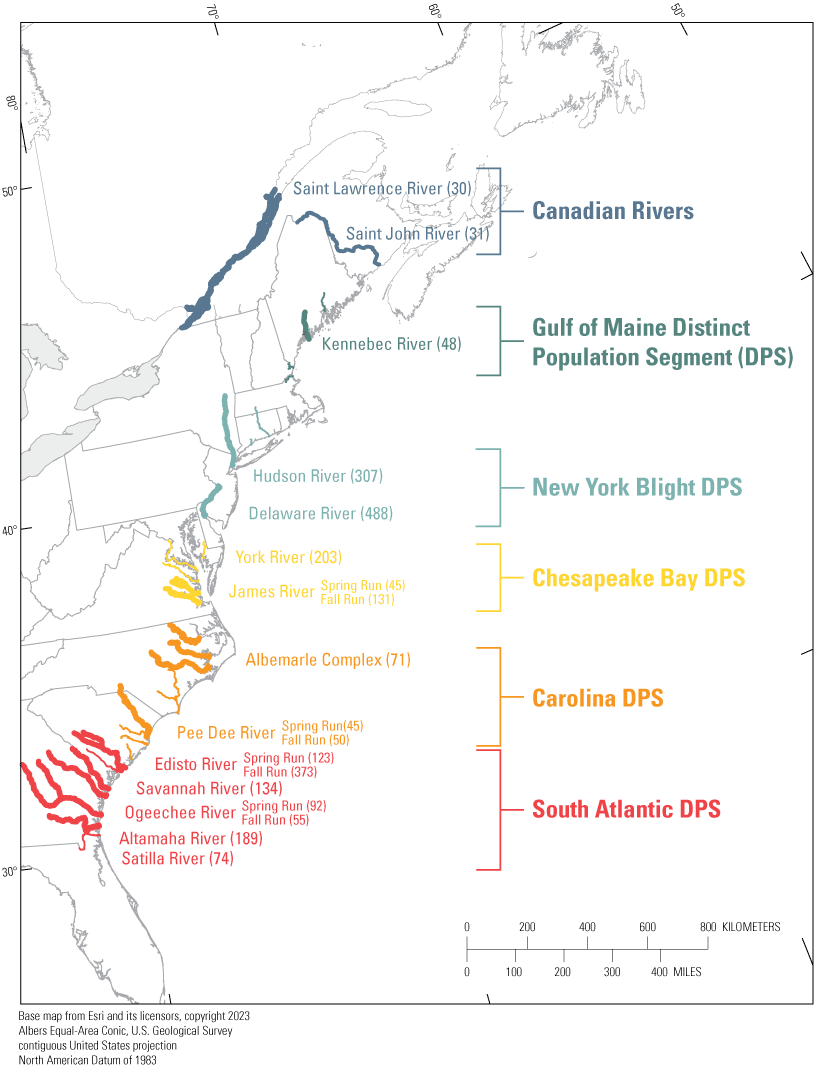
Distribution of 18 Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) collections in the genetic baseline that served as reference populations for individual-based assignment tests of unknown individuals. Collections are color-coded by distinct population segment (DPS). The sample size of individuals from each collection appears in parentheses. Figure modified from White and others (2021).
Most of the individuals in the baseline were genotyped at USGS-EESC; however, some samples from the James and Edisto rivers were genotyped by the South Carolina Department of Natural Resources (SCDNR) using similar methods, with the exception that SCDNR did not genotype at locus LS39. A previous effort to standardize allele calls between USGS-EESC and SCDNR found very strong concordance between the two laboratories. However, there were occasional differences at Aox12 locus that pertained to the genotype 187|189. As a result, the SCDNR genotype 187|189 for locus Aox12 was omitted and the locus treated as missing data. This omission has negligible influence on the genetic baseline, as it only affected two individuals from the Pee Dee Fall Run and three individuals from James Fall Run.
The majority of individuals in the genetic baseline are river-resident juveniles (less than 500 millimeters [mm] total length [TL]) and adults (greater than 1,500 mm TL). Individuals outside of these size criteria were excluded from the genetic baseline due to the extensive dispersal of subadult and adult Atlantic sturgeon outside of spawning season. The only exceptions were two individuals from the Pee Dee River and seven individuals from the St. John River that were 1,200–1,400 mm TL, which were included due to limited sample sizes in those locations and because the sturgeon were captured at a time and locality consistent with spawning. With the exception of the sturgeon representing the Albemarle Sound, all individuals included in the baseline were captured in riverine environments. Albemarle Sound samples were collected in tidal waters outside of a known spawning area. The inclusion of individuals collected outside of putative spawning reaches in the Albemarle Sound is a known weakness of the current baseline, as the collection likely represents sturgeon from several spawning populations within the Carolina DPS.
White and others (2021) demonstrated high assignment accuracy of the genetic baseline with 81.7 percent of individuals assigning to the correct river of origin and 94.0 percent of individuals assigning to the correct DPS of origin (table 1). High assignment accuracy suggests that the baseline has high sensitivity and specificity and can be used to reliably assign individuals of unknown origin to a population or DPS of origin.
Table 1.
Classification confusion matrix for individual based assignments using GeneClass2 for 18 collections of Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) used in the baseline dataset.[Table from White and others (2021). n, number of samples; St., Saint; —, no data; DPS, distinct population segment]
Individual-based genetic assignments on 345 unknown samples provided by NMFS were conducted in the program GeneClass2 (Piry and others. 2004) using the criterion of Rannala and Mountain (1997). Individuals were assigned to each of the 18 reference collections in the baseline using two methods. In the first, the negative-log likelihood of assignment to each collection was used to identify the five most probable populations of origin. Assignment strength to each of these populations was evaluated by calculating a score using equation 1
whereis the likelihood that individual (i) is natal to population (l), and
is the sum of all likelihood values across the 18 collections.
Although likelihood scores indicate the most likely source among the candidate populations, they do not provide information as to whether or not a candidate population could reasonably have produced a particular individual (in other words, goodness-of-fit). To provide this additional perspective, assignments were also completed using Monte-Carlo resampling to calculate the probability that each multi-locus genotype could be drawn from each reference population based on the allele frequencies in that reference population.
However, one caution is that assignments are based solely on allele frequencies, and it is possible (although unlikely) that by chance an individual has a genotype that is more common in another population. Misassignment happens most frequently when individuals are scored at fewer than 12 loci or when populations have similar allele frequencies—which frequently occurs among populations within the same DPS. It is also important to note that the assignment testing approach used in this analysis requires an individual to be assigned to one of the 18 reference collections. In other words, individuals could not be assigned to an extant population that was not represented in the genetic baseline, even if that was its true source.
Results
The average number of loci genotyped across the 345 samples was 11.3 (range: 0–12; White and others, 2023). Sixteen individuals were genotyped at fewer than four loci and were removed from further analysis. In addition, one pair of identical multi-locus genotypes was identified (Aoxy30995 and Aoxy30996). As the probability of two randomly sampled individuals having identical genotypes is less than 0.0001 percent, the repeated genotype was assumed to represent a duplicate sample from the same individual and one was removed from the analysis. Thus, there were 329 individuals available for assignment, of which 116 samples included geographic coordinates for the sample location, and all were collected between Cape May, New Jersey and Virginia Beach, Virginia. Collection location for the remaining 213 samples was not provided to USGS; however, those individuals were included in all analyses.
The majority of samples assigned to the Hudson River (130), Delaware River (50), James River Fall Run (42), York River (21), and Pee Dee River Fall Run (15) populations. Less than 5 percent of individuals assigned to the Kennebec River, Ogeechee River Spring Run, Albemarle Complex, Savannah River, Pee Dee River Spring Run, St. John River, Altamaha River, Edisto River Spring Run, Satilla River, Ogeechee River Fall Run, James River Spring Run, and St. Lawrence River populations (fig. 2; table 2). Most of these assignments were made with a high degree of likelihood (mean score: 91.0 percent to the assigned population; range: 37.5–100.0 percent). Of the 90 individuals with a score less than 90 percent, the second most probable population was in the same DPS 51 percent of the time (White and others, 2023).
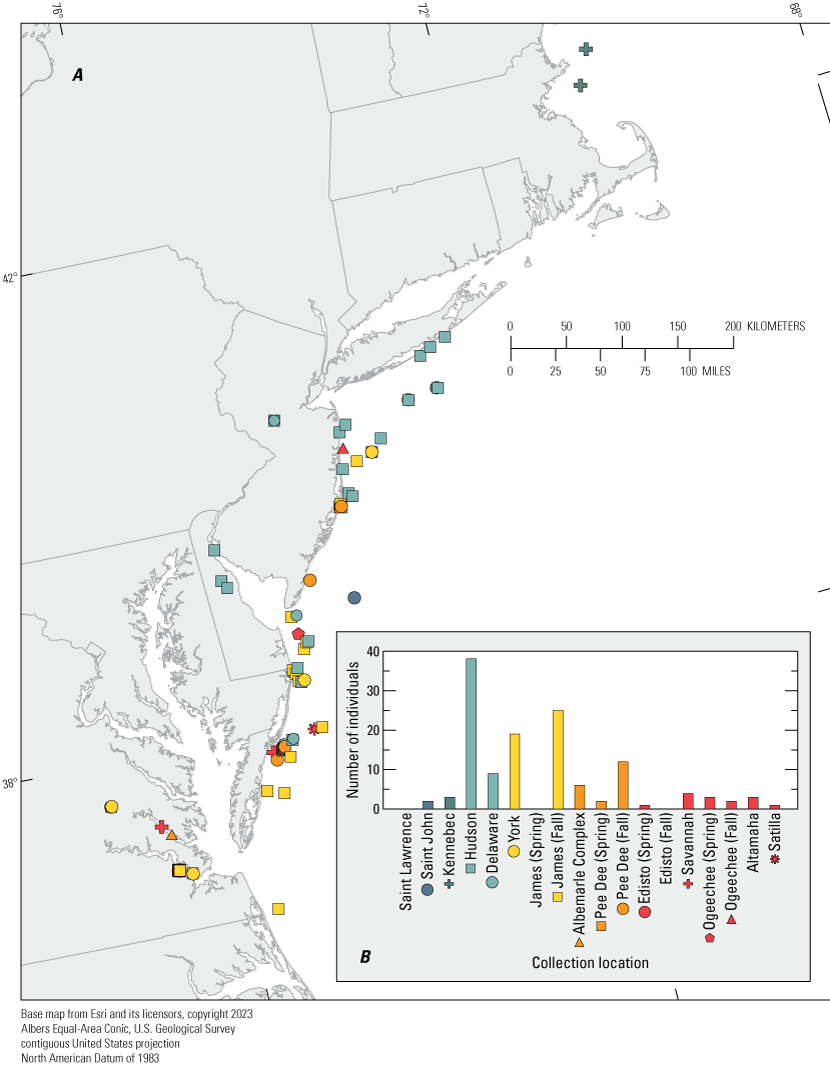
Results of genetic assignment tests for 116 Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) with reported collection locations along the Atlantic Coast of North America. A, Map showing individual collection locations, symbolized by results of population assignment tests. B, Bar graph showing number of individuals assigned to each population for the 116 individuals where capture location is known.
Table 2.
Number of Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) assigned to each of the 18 collections in the genetic baseline.[DPS, distinct population segment; St., Saint; —, no data]
There was a reasonable probability that most individuals could have originated in their assigned populations given reference allele frequencies (mean: 41.5 percent; range: 0.00–99.9 percent). There were nine individuals that assigned with 0.00 percent probability to all collections, which included four individuals that assigned to the James River Fall Run, one to Ogeechee River Spring Run, and three to Pee Dee River Spring Run using the likelihood-based method; however, these individuals were retained in the analyses (White and others, 2023). These individuals may have possessed allele combinations which were not present in the baseline, or alternatively may have originated in populations that are not represented in the baseline.
Of the 272 individuals that were screened for sex, there were 121 females, 133 males, and 18 unknowns (13 of these unknowns had intermediate RFU values and a further 5 did not amplify at control loci). Two individuals that were categorized as unknown also failed to be genotyped at a sufficient number of loci for population assignment testing and were removed from the analysis as was the presumed repeated sample (thus leaving a total sample size for sex determination of 269 individuals). The number of males and females assigned to each population was approximately equal (table 3).
Table 3.
Molecular sex identification for 269 Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon) which were assigned to collections in the genetic baseline.[St., Saint; —, no data]
Summary
In a sample of 329 Acipenser oxyrinchus oxyrinchus (Atlantic sturgeon), we assigned individuals to 17 of 18 collections described in the genetic baseline by White and others, (2021). Nearly 40 percent of individuals assigned to the Hudson River population, which is unsurprising given that the population is believed to be one of the most numerically abundant (Breece and others, 2021). Moreover, all individuals with reported sample locations were collected in the mid-Atlantic region, which is in close proximity to the mouth of the Hudson River.
The only collection in the baseline that was not detected in the sample of unknown individuals was that of the Edisto River Fall Run; however, individuals from the Edisto River Spring Run were detected. Furthermore, 10 percent of all individuals were assigned to populations in the South Atlantic distinct population segment (DPS). This finding supports previous research which noted the propensity for long-distance migrations in subadult and adult Atlantic sturgeon, including the potential for populations from the South Atlantic DPS to be encountered at more northern latitudes (Kazyak and others, 2021). Additionally, the mid-Atlantic region has been implicated as an area of extensive stock mixing with populations from all five DPS and the Canadian Rivers frequently detected in this region (Waldman and others, 2013; O’Leary and others, 2014; Wirgin and others 2015; Rulifson and others, 2020).
Although sample location was not provided for more than half of the individuals, and most samples were collected in offshore environments, it is notable that many Atlantic sturgeon collected in riverine and estuarine environments in Virginia were assigned to populations located outside the Chesapeake Bay DPS. This, along with the finding of long-distance migrations, highlights the potential for local management activities to effect Atlantic sturgeon populations from distance sources.
It is notable that the individuals that we genetically sexed in our analyses represented a nearly equal sex ratio with 121 females and 132 males. Although this result may be potentially biased by sample location and method, it does suggest that adult sex ratio in oceanic environments may be relatively even. The even sex ratio is in opposition to what is commonly reported in riverine environments, where samples are often dominated by males (Hilton and others, 2016). Differences in sex ratio between marine and freshwater habitats may represent differences in behavior, with females returning less frequently in riverine environments (and thus less available for sampling) or sex-specific patterns of mortality.
References Cited
Atlantic States Marine Fisheries Commission, 1998, Amendment 1 to the Interstate Fishery Management Plan for Atlantic sturgeon: Fishery Management Report 31, prepared by Atlantic States Marine Fisheries Commission and Atlantic Sturgeon Plan Development Team Washington D.C., accessed December 25, 2021, at http://www.asmfc.org/uploads/file/sturgeonAmendment1.pdf.
Atlantic States Marine Fisheries Commission, 2017, 2017 Atlantic sturgeon benchmark stock assessment and peer review report, Washington, D.C., accessed December 25, 2021, at http://www.asmfc.org/uploads/file//59f8d5ebAtlSturgeonBenchmarkStockAssmt_PeerReviewReport_2017.pdf.
Atlantic Sturgeon Status Review Team, 2007, Status review of Atlantic sturgeon (Acipenser oxyrinchus oxyrinchus): National Marine Fisheries Service, Northeast Regional Office, accessed December 25, 2021, at https://repository.library.noaa.gov/view/noaa/16197.
Breece, M.W., Higgs, A.L., and Fox, D.A., 2021, Spawning intervals, timing, and riverine habitat use of adult Atlantic sturgeon in the Hudson River: Transactions of the American Fisheries Society, v. 150, no. 4, p. 528–537, accessed December 25, 2021, at https://afspubs.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/tafs.10304.
Committee on the Status of Endangered Wildlife in Canada, 2011, COSEWIC assessment and status report on the Atlantic sturgeon Acipenser oxyrinchus in Canada: Committee on the Status of Endangered Wildlife in Canada, Ottawa, accessed December 25, 2021, at https://publications.gc.ca/collections/collection_2012/ec/CW69-14-636-2011-eng.pdf.
Dadswell, M.J., 2006, A review of the status of Atlantic sturgeon in Canada, with comparisons to populations in the United States and Europe: Fisheries (Bethesda, Md.), v. 31, no. 5, p. 218–229, accessed December 25, 2021, at https://doi.org/10.1577/1548-8446(2006)31[218:AROTSO]2.0.CO;2.
Dadswell, M.J., Wehrell, S.A., Spares, A.D., Mclean, M.F., Beardsall, J.W., Logan‐Chesney, L.M., Nau, G.S., Ceapa, C., Redden, A.M., and Stokesbury, M.J., 2016, The annual marine feeding aggregation of Atlantic sturgeon Acipenser oxyrinchus in the inner Bay of Fundy—Population characteristics and movement: Journal of Fish Biology, v. 89, no. 4, p. 2107–2132, accessed December 25, 2021, at https://doi.org/10.1111/jfb.13120.
Dunton, K.J., Jordaan, A., Conover, D.O., McKown, K.A., Bonacci, L.A., and Frisk, M.G., 2015, Marine distribution and habitat use of Atlantic sturgeon in New York lead to fisheries interactions and bycatch: Marine and Coastal Fisheries, v. 7, no. 1, p. 18–32, accessed June 30, 2022, at https://doi.org/10.1080/19425120.2014.986348.
Dunton, K.J., Jordaan, A., McKown, K.A., Conover, D.O., and Frisk, M.G., 2010, Abundance and distribution of Atlantic sturgeon (Acipenser oxyrinchus) within the northwest Atlantic Ocean, determined from five fishery-independent surveys: Fish Bulletin, v. 108, no. 4, p. 450–465, accessed December 25, 2021, at https://spo.nmfs.noaa.gov/sites/default/files/pdf-content/2010/1084/dunton.pdf.
Erickson, D.L., Kahnle, A., Millard, M.J., Mora, E.A., Bryja, M., Higgs, A., Mohler, J., DuFour, M., Kenney, G., Sweka, J., and Pikitch, E.K., 2011, Use of pop-up satellite archival tags to identify oceanic-migratory patterns for adult Atlantic sturgeon, Acipenser oxyrinchus oxyrinchus Mitchell, 1815: Journal of Applied Ichthyology, v. 27, no. 2, p. 356–365, accessed December 25, 2021, at https://doi.org/10.1111/j.1439-0426.2011.01690.x.
Fox, A.G., Wirgin, I.I., and Peterson, D.L., 2018, Occurrence of Atlantic sturgeon in the St. Marys River, Georgia: Marine and Coastal Fisheries, v. 10, no. 6, p. 606–618, accessed December 25, 2021, at https://doi.org/10.1002/mcf2.10056.
Henderson-Arzapalo, A., and King, T.L., 2002, Novel microsatellite markers for Atlantic sturgeon (Acipenser oxyrhinchus) population delineation and broodstock management: Molecular Ecology Notes, v. 2, no. 4, p. 437–439, accessed December 25, 2021, at https://doi.org/10.1046/j.1471-8286.2002.00262.x.
Hilton, E.J., Kynard, B., Balazik, M.T., Horodysky, A.Z., and Dillman, C.B., 2016, Review of the biology, fisheries, and conservation status of the Atlantic sturgeon, (Acipenser oxyrinchus oxyrinchus Mitchill, 1815): Journal of Applied Ichthyology, v. 32, no. 1, p. 30–66, accessed December 25, 2021, at https://doi.org/10.1111/jai.13242.
Kazyak, D.C., Flowers, A.M., Hostetter, N.J., Madsen, J.A., Breece, M., Higgs, A., Brown, L.M., Royle, J.A., and Fox, D.A., 2020, Integrating side-scan sonar and acoustic telemetry to estimate the annual spawning run size of Atlantic sturgeon in the Hudson River: Canadian Journal of Fisheries and Aquatic Sciences, v. 77, no. 6, p. 1038–1048, accessed December 25, 2021, at https://doi.org/10.1139/cjfas-2019-0398.
Kazyak, D.C., White, S.L., Lubinski, B.A., Johnson, R., and Eackles, M., 2021, Stock composition of Atlantic sturgeon (Acipenser oxyrinchus oxyrinchus) encountered in marine and estuarine environments on the U.S. Atlantic Coast: Conservation Genetics, v. 22, p. 767–781, accessed December 25, 2021, at https://doi.org/10.1007/s10592-021-01361-2.
King, T.L., Lubinski, B.A., and Spidle, A.P., 2001, Microsatellite DNA variation in Atlantic sturgeon (Acipenser oxyrinchus oxyrinchus) and cross-species amplification in the Acipenseridae: Conservation Genetics, v. 2, p. 103–119, accessed December 25, 2021, at https://doi.org/10.1023/A:1011895429669.
Kuhl, H., Guiguen, Y., Höhne, C., Kreuz, E., Du, K., Klopp, C., Lopez-Roques, C., Yebra-Pimentel, E.S., Ciorpac, M., Gessner, J., and Holostenco, D., 2021, A 180 Myr-old female-specific genome region in sturgeon reveals the oldest known vertebrate sex determining system with undifferentiated sex chromosomes: Philosophical Transactions of the Royal Society B, v. 376, no. 1832, 10 p., accessed June 18, 2022, at https://doi.org/10.1098/rstb.2020.0089.
May, B., Krueger, C.C., and Kincaid, H.L., 1997, Genetic variation at microsatellite loci in sturgeon—Primer sequence homology in Acipenser and Scaphirhynchus: Canadian Journal of Fisheries and Aquatic Sciences, v. 54, no. 7, p. 1542–1547, accessed December 25, 2021, at https://doi.org/10.1139/f97-061.
National Oceanic and Atmospheric Administration, 2012a, Endangered and threatened wildlife and plants; threatened and endangered status for distinct population segments of Atlantic sturgeon in the northeast region: Federal Register, v. 77, p. 5880–5912, accessed December 27, 2021, at https://www.govinfo.gov/content/pkg/FR-2012-02-06/pdf/2012-1946.pdf.
National Oceanic and Atmospheric Administration, 2012b, Endangered and threatened wildlife and plants; final listing determinations for two distinct population segments of Atlantic sturgeon (Acipenser oxyrinchus oxyrinchus) in the southeast: Federal Register, v. 77, p. 5914–5982, accessed December 27, 2021, at https://www.govinfo.gov/content/pkg/FR-2012-02-06/pdf/2012-1950.pdf.
O’Leary, S.J., Dunton, K.J., King, T.L., Frisk, M.G., and Chapman, D.D., 2014, Genetic diversity and effective size of Atlantic sturgeon, Acipenser oxyrhinchus oxyrhinchus river spawning populations estimated from the microsatellite genotypes of marine-captured juveniles: Conservation Genetics, v. 15, no. 5, p. 1173–1181, accessed December 25, 2021, at https://doi.org/10.1007/s10592-014-0609-9.
Piry, S., Alapetite, A., Cornuet, J.M., Paetkau, D., Baudouin, L., and Estoup, A., 2004, GeneClass2—A software for genetic assignment and first-generation migrant detection: The Journal of Heredity, v. 95, no. 6, p. 536–539, accessed December 25, 2021, at https://doi.org/10.1093/jhered/esh074.
Rannala, B., and Mountain, J.L., 1997, Detecting immigration by using multilocus genotypes: Proceedings of the National Academy of Sciences of the United States of America, v. 94, no. 17, p. 9197–9201, accessed December 25, 2021, at https://doi.org/10.1073/pnas.94.17.9197.
Rulifson, R.A., Bangley, C.W., Cudney, J.L., Dell’Apa, A., Dunton, K.J., Frisk, M.G., Loeffler, M.S., Balazik, M.T., Hager, C., Savoy, T., Brundage, H.M., III, and Post, W.C., 2020, Seasonal presence of Atlantic sturgeon and sharks at Cape Hatteras, a large continental shelf constriction to coastal migration: Marine and Coastal Fisheries, v. 12, no. 5, p. 308–321, accessed December 25, 2021, at https://doi.org/10.1002/mcf2.10111.
Waldman, J.R., King, T., Savoy, T., Maceda, L., Grunwald, C., and Wirgin, I., 2013, Stock origins of subadult and adult Atlantic sturgeon, Acipenser oxyrinchus, in a non-natal estuary, Long Island Sound: Estuaries and Coasts, v. 36, no. 2, p. 257–267, accessed December 25, 2021, at https://doi.org/10.1007/s12237-012-9573-0.
White, S.L., Kazyak, D.C., Darden, T.L., Farrae, D.J., Lubinski, B.A., Johnson, R.L., Eackles, M.S., Balazik, M.T., Brundage, H.M., III, Fox, A.G., Fox, D.A., Hager, C.H., Kahn, J.E., and Wirgin, I.I., 2021, Establishment of a microsatellite genetic baseline for North American Atlantic sturgeon (Acipenser o. oxyrhinchus) and range-wide analysis of population genetics: Conservation Genetics, v. 22, no. 6, p. 977–992, accessed December 27, 2021, at https://doi.org/10.1007/s10592-021-01390-x.
White, S.L., Lubinski, B.A., Johnson, R., Eackles, M., and Kazyak, D.C., 2023, Individual assignments and microsatellite genotypes for Atlantic sturgeon from 2021: U.S. Geological Survey data release, at https://doi.org/10.5066/P943B0GT.
Wirgin, I., Maceda, L., Grunwald, C., and King, T., 2015, Population origin of Atlantic sturgeon Acipenser oxyrinchus oxyrinchus by-caught in U.S. Atlantic coast fisheries: Journal of Fish Biology, v. 86, no. 4, p. 1251–1270, accessed December 25, 2021, at https://doi.org/10.1111/jfb.12631.
For more information, contact
Director, Eastern Ecological Science Center
11649 Leetown Road,
Kearneysville, WV 25430
Or visit our website at https://www.usgs.gov/centers/eesc.
Publishing support provided by the Baltimore Publishing Service Center.
Disclaimers
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Although this information product, for the most part, is in the public domain, it also may contain copyrighted materials as noted in the text. Permission to reproduce copyrighted items must be secured from the copyright owner.
Suggested Citation
White, S.L., Johnson, R.L., Lubinski, B.A., Eackles, M.S., and Kazyak, D.C., 2023, Genetic population assignments of Atlantic sturgeon provided to National Marine Fisheries Service, 2022: U.S. Geological Survey Open-File Report 2023–1054, 10 p., https://doi.org/10.3133/ofr20231054.
ISSN: 2331-1258 (online)
Study Area
| Publication type | Report |
|---|---|
| Publication Subtype | USGS Numbered Series |
| Title | Genetic population assignments of Atlantic sturgeon provided to National Marine Fisheries Service, 2022 |
| Series title | Open-File Report |
| Series number | 2023-1054 |
| DOI | 10.3133/ofr20231054 |
| Publication Date | August 24, 2023 |
| Year Published | 2023 |
| Language | English |
| Publisher | U.S. Geological Survey |
| Publisher location | Reston, VA |
| Contributing office(s) | Eastern Ecological Science Center |
| Description | Report: vi, 10 p.; Data Release |
| Country | Canada, United States |
| Online Only (Y/N) | Y |
| Additional Online Files (Y/N) | N |


