Proceedings of the 2024 Asia-Pacific Wildlife Health Workshop—Collaborating Against Shared Threats
Links
- Document: Report (2.4 MB pdf) , HTML , XML
- Download citation as: RIS | Dublin Core
Preface
Emerging diseases of wildlife origin are increasingly transboundary (they spread rapidly across geographic regions and across continents). In recent years, examples include the rapid spread of African swine fever across Europe and Asia with negative effects on food security, and the near global spread of highly pathogenic avian influenza which has devastated wildlife populations, caused economic harm, and which threatens public health; consequently, international partnerships and networks are essential to facilitate the sharing of information for improved situational awareness and better preparedness and response. In this regard, the U.S. Geological Survey and the Korea National Institute for Wildlife Disease Control and Prevention have had a long-standing partnership to foster scientific collaboration. A key part of the activities has been annual scientific workshops, which commenced in 2016.
The 2024 workshop in Hilo, Hawaii, (appendix 1) was the most recent in these series of workshops and included participants from across Asia and the Pacific region, including Thailand, Vietnam, China, Republic of Korea, Japan, Australia, Cook Islands, Fiji, and the United States. The goals of the workshop were:
-
• to continue to build the wildlife health community of practice in the Asia-Pacific region and expand the participants to agencies and institutions from other countries in the region; and
-
• exchange scientific knowledge among the participants to share best practices, create scientific networks, and build capacity in wildlife health science for the Asia-Pacific region.
The themes discussed at the workshop included wildlife health risk management, avian Influenza, African swine fever, climate change and emerging diseases, and international cooperation. This report contains the author-submitted abstracts which provide a summary of the presentations and discussions during the workshop. The aim is to share this information to continue to foster international scientific exchange to protect wildlife health, livestock, and public health from the negative impacts of infectious and noninfectious diseases.
Introducing the Wildlife Health Australia Collaborating Centre in Wildlife Health Risk Management—Working Regionally in the Interests of Australia’s Biosecurity and Biodiversity Resilience
Steve Unwin1 and Erin Davis1
1Wildlife Health Australia.
Abstract
Despite global pandemics and epidemics in multiple species, from severe acute respiratory syndrome coronavirus 2 to multi-drug resistant mycobacterium tuberculosis complex to malaria to ASF to highly pathogenic avian influenza (HPAI), our species remains resistant to taking a holistic view of health. Pathogens do not recognize geopolitical borders. In the Anthropocene, human activity is increasing the number and size of interfaces between us and other animal species. When the One Health High Level Expert Panel developed a new definition for One Health (Adisasmito and others, 2022), incorporating the need to better manage the risks through coordination, collaborating, communicating and capacity building, an opportunity to take a more prevention-focused systems thinking approach emerged. Figure 1 highlights the various scales we need to consider in wildlife health risk management at a systems level, supported by the One Health High Level Expert Panel approach to One Health.
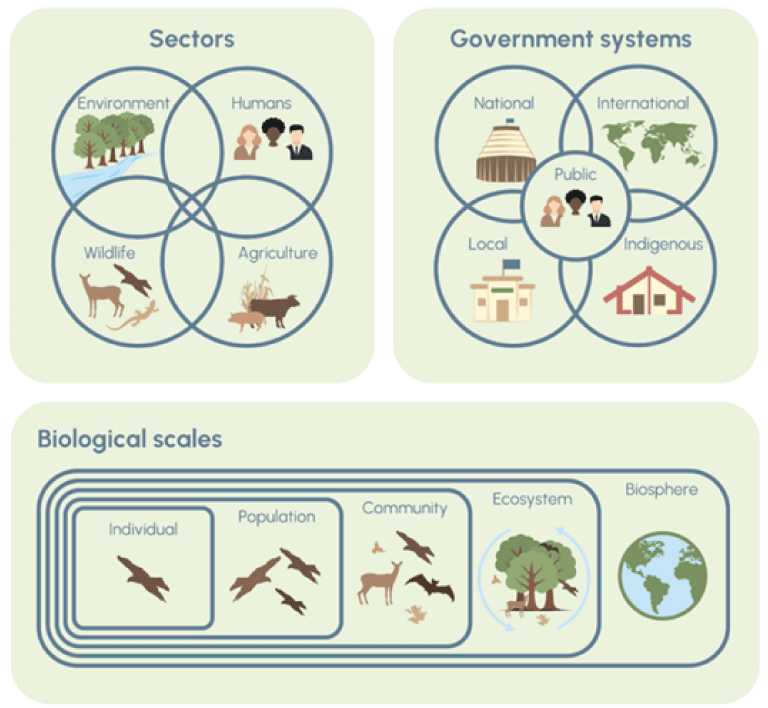
Diagrams showing that an understanding of a systems approach is critical in the Anthropocene for effective One Health (Pepin and others, 2024).
Understanding this dilemma, in 2023 Wildlife Health Australia (WHA)became home to a World Organisation for Animal Health (WOAH) Collaborating Centre in Wildlife Health Risk Management to help others in the region grow capacity and capability in wildlife health. We link our activities to the WOAH wildlife health framework outcomes and outputs and have three main objectives.Objective 1: Successful integration of wildlife health into One Health decision-making.Objective 2: Improving prevention and mitigation capabilities for current and emerging health risks, including those with pandemic potential, through interdisciplinary collaboration.
In the Asia Pacific, WHA and our partners are taking an all-hazards global systems approach to help identify and fill data gaps to improve surveillance outcomes.
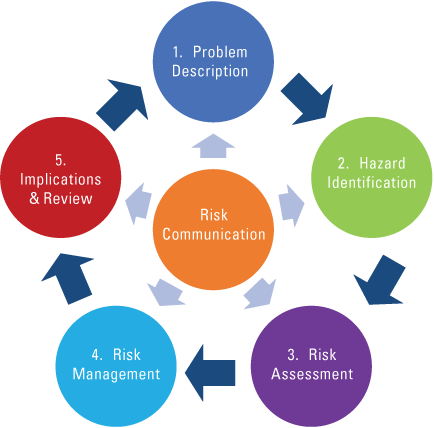
Diagram showing the wildlife disease risk analysis process (Jakob-Hoff and others, 2014).
An example is our partnership with the International Union for Conservation of Nature Conservation Planning Specialist Group to build capacity in Wildlife Disease Risk Analysis (WDRA). This is a process and a tool to assist with contextualizing and managing the health of wildlife. Figure 2 highlights how the process is iterative. A second edition of the WDRA guidelines is in preparation, broadening the remit from disease exclusively to analyzing risks to wildlife health more broadly, in line with a systems all-hazard approach. As scientists, we are provided skills to explain the world around us that can help society understand, care about, and manage how we work within it. As humans, we are the story-telling apes attempting to explain the world around us. When faced with health or disease challenges, WDRA is simply a way of thinking matched with an objective tool. It helps us to describe and assess a situation, and as an applied tool, it helps inform how we should manage, implement, and review or evaluate what we do, all focused on successful communications links. So, it provides pathways to solutions scientifically and tells the story in a way that should be easily understood. This process and online course are now embedded within the collaborating center activities.Objective 3: Establishing a sustainable cadre of professionals able to identify and manage drivers of emerging health risks.
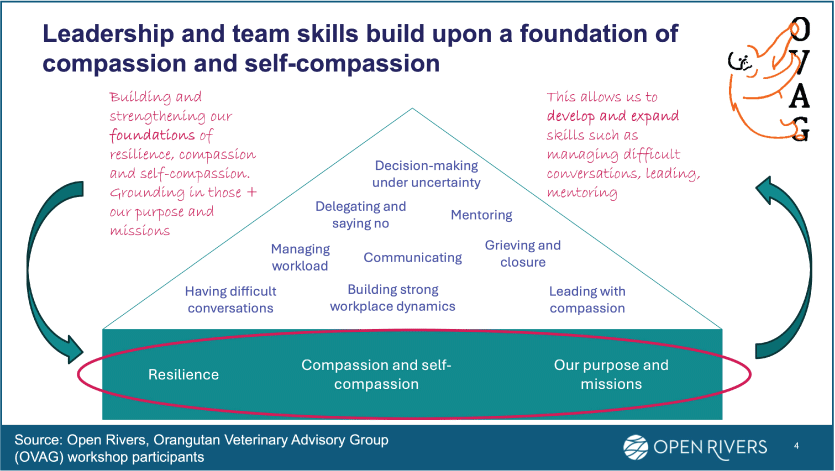
Diagram showing how leadership and team skills build on a foundation of compassion and self-compassion.
To achieve objective 3, WHA realized we needed to understand how professional communities of practice can be sustained. Figure 3 presents a framework from WHA partners Orangutan Veterinary Advisory Group, a network of Asian ape specialists concentrated in Indonesia and Malaysia formed in 2009, and Open Rivers Consultants. It illustrates a bespoke resilience program generated by the participants in this wildlife health community of practice to help facilitate network participant resilience.
To achieve successful outcomes for biodiversity and biosecurity, WHA promotes and participates in better systems management (objectives 1 and 2), supporting inclusive and sustainable networks (objective 3).
Animal Health Capacity Building—The Role of Para-Vets in Animal Biosecurity for the Pacific Region
Elenoa Salele1*
1Pacific Community.
*Correspondence: elenoas@spc.int
Abstract
Livestock play a significant role in the cultures of Pacific Island countries and territories and are important to their economies and food security. Animal production services are under-resourced across the Pacific and veterinary services are sparse with livestock farmers in many countries having limited access to veterinary services. Despite the prominence of livestock, the region experiences various challenges in sustaining animal health and production.
At the First Regional Conference of Ministers of Agriculture and Forestry Services organized by the Pacific Community (SPC) in September 2004, the ministers acknowledged the need to strengthen livestock production systems and the growing threats from exotic diseases and further recommended the establishment of the Pacific Heads of Veterinary and Animal Production Services as a mechanism to facilitate regional collaboration in developing programs to strengthen animal health and production services across the Pacific.
The SPC developed the Pacific Animal Health and Production Framework in response to the Pacific Heads of Agriculture and Forestry Services meeting in 2019 that directed the SPC to urgently address the shortages of veterinary services and threats posed by zoonotic diseases to the livestock industry as well as human health.
Its development involved an iterative process through various consultations with stakeholders in the livestock sector from the region and in countries. The process also involved an extensive literature review of past and ongoing livestock research and development activities as well as national policies on animal agriculture.
The shortage of qualified veterinarians and limited access to veterinary services negatively impacts biosecurity planning, animal health, and production, and the overall safeguarding of veterinary public health was identified as the top challenging priority from SPC member countries.
In response to this pressing need, SPC developed a para-veterinarian (para-vet) training program to address these gaps and enhance the region's animal health capacity. Para-vets serve as the frontline support for veterinarians at the community or field level, providing essential animal healthcare and production guidance to livestock farmers in their respective communities. This was formalized through a memorandum of understanding between SPC, Agricultural Development in the American Pacific, and the University of South Pacific in 1999. In this memorandum, the three institutions agreed to collaborate on the development and dissemination of the PARAVET training program. Since its inception, SPC’s para-vet training has been conducted across 15 Pacific Island Countries and Territories, resulting in a total of over 400 graduates. The training program receives technical support from the Australian Department of Agriculture, Fisheries and Forestry through the Pacific Engagement Program for Animal Health team, as well as the New Zealand Ministry of Primary Industry (MPI) through the animal health and biosecurity team.
In October 2022, the SPC–European Union funded project “SAFE Pacific Project” in collaboration with the Australian Department of Agriculture, Fisheries and Forestry conducted training on para-vet animal disease and surveillance in partnership with the Solomon Islands Ministry of Agriculture and Livestock (MAL).
The effectiveness of this surveillance training came to the forefront as livestock officers from MAL received reports of mass chicken deaths on farms, resulting in the detection of a virus responsible for high mortality rates of chicks in Guadalcanal, Solomon Islands, in July 2023.
Knowledge acquired from the previous para-vet training was successfully applied by the MAL officers as they were able to conduct surveillance and monitoring activities in Solomon Islands. Upon receiving reports of mass mortalities in chicken flocks in July 2023, MAL officers utilized their enhanced capabilities to visit different farms and obtain appropriate samples, which they sent to the Australian Centre for Disease Preparedness for a confirmatory diagnosis.
To conclude, acknowledging the logistical and other challenges associated with carrying out wildlife work in Pacific Island countries, there is an opportunity to apply para-vet training to wildlife-related work, to help enhance biosecurity, and help improve One Health outcomes in these regions.
Keywords: SPC, para-vet, livestock, animal biosecurity, Pacific Islands
Collaborations Between Biologists and Veterinarians Yield Insights into Marine Turtle Fibropapillomatosis
Thierry M. Work1* and George H. Balazs2
1U.S. Geological Survey.
2Golden Honu Services of Oceania.
*Correspondence: Thierry_work@usgs.gov
Abstract
Fibropapillomatosis (FP) is a tumor disease affecting mainly green turtles (Chelonia mydas) throughout their range globally. FP was first documented in green turtles in Florida (United States) in the early 1930s and subsequent investigations in that state described the gross and microscopic pathology of the disease showing that it was infections and likely associated with a herpes virus. FP appeared in Hawaiian green turtles in the 1950s but did not reach epizootic magnitudes until the early 1990s. Collaborative investigations between the National Oceanic and Atmospheric Administration (NOAA) and the U.S. Geological Survey (USGS) between the mid-1990s to mid-2000s have yielded substantial insights into FP. Specifically, the virus associated with FP was identified as chelonid alphaherpesvirus 5 using newly developed molecular tools. Long-term studies showed FP followed a classic epidemic curve indicating drivers of the disease are diminishing in Hawaii. We showed that FP led to immunosuppression and secondary bacterial infections and demonstrated that disease in green turtle manifests differently in Hawaii versus Florida. Transmission of the virus in the wild is likely through shedding from tumors or transmission by leeches or cleaner wrasse. Finally, tools were developed to grow chelonid alphaherpesvirus 5 in the lab using a three-dimensional tissue culture. None of these advances would have been possible without a robust statewide stranding program managed by NOAA that allowed access to specimens for research. This highlights the importance of cross-agency collaborations to address complex wildlife health problems in challenging settings (marine environment).
Keywords: green turtle, tumors, epizootic, virology
Mitigating Transboundary Emerging Infectious Diseases—Wildlife and One Health Sustainability
Mathuros Tipayamongkholgul,1 Cheerawit Rattanapan,1 Christine Stanly,1 Walasinee Sakcamduang,1 Sarin Suwanpakdee,1* and Anuwat Wiratsudakul1*
1Mahidol University.
*Correspondence: sarin.suw@mahidol.edu; anuwat.wir@mahidol.edu
Abstract
The Mahidol University Global Health Consultative Meeting, a 2-day consultative meeting held in March 2024 at Mahidol University, Thailand, brought together 25 experts to tackle the growing threat of transboundary emerging infectious diseases (TEIDs) in Southeast Asia. Employing a One Health approach, the meeting aimed to unite human and animal health sectors to combat diseases like AI, rabies, and coronaviruses. Discussions focused on developing a comprehensive strategy against TEIDs. This strategy includes strengthening veterinary services, improving disease surveillance, fostering joint research, and harmonizing border control measures. A key recommendation was the establishment of an Association of Southeast Asian Nations (ASEAN) One Health Collaborative Network to facilitate information exchange and collaboration. This network, along with a proposed wildlife disease diagnostic laboratory network, would enable rapid and standardized testing, improving wildlife health surveillance. Recognizing the importance of skilled personnel, the meeting emphasized capacity building for wildlife disease response. Training programs for field workers, healthcare professionals, and veterinarians were crucial. Sharing best practices and innovative methods for wildlife conservation across ASEAN member countries was also highlighted. Standardization of procedures for surveillance and laboratory analysis was identified as essential for timely and accurate data collection. Additionally, promoting health literacy, particularly in wildlife farming, was seen as vital to preventing zoonotic disease transmission by raising awareness of One Health principles. The meeting concluded that sustainable management of biodiversity and environmental health is critical for mitigating TEIDs. A united ASEAN front, supported by the proposed collaborative network, would promote regional cooperation and sustainable practices aligned with the Sustainable Development Goals. This multifaceted approach promises a healthier future for humans, animals, and the environment. By fostering collaboration, innovation, and a commitment to One Health, ASEAN can effectively address the complexities of TEIDs. The meeting's outcomes provide a strong foundation for future initiatives and underscore the need for continued regional efforts.
Keywords: transboundary emerging infectious diseases, One Health, ASEAN, wildlife health surveillance, zoonotic diseases
System-Wide Approach to Wildlife Health Capacity Enhancement in Southeast Asia
Sarin Suwanpakdee,1 Nareerat Sangkachai,1 Anuwat Wiratsudakul,1 Witthawat Wiriyarat,1 Walasinee Sakcamduang,1 Peerawat Wongluechai,1 Choenkwan Pabutta,1 Ladawan Sariya,1 Waruja Korkijthamkul,1 David S. Blehert,2 C. LeAnn White,2 Daniel P. Walsh,2 Craig Stephen,3 Parntep Ratanakorn,1 and Jonathan M. Sleeman1,2*
1Mahidol University.
2U.S. Geological Survey.
3McEachran Institute.
*Correspondence: jsleeman@usgs.gov; 608-280-1135
Abstract
There is an increasing need for robust wildlife health programs that provide surveillance and management for diseases in wildlife and wild aquatic populations. This paper illustrates the value of a systematic method to enhancing wildlife health programs using a case study. The USGS and Mahidol University, Faculty of Veterinary Science, Thailand National Wildlife Health Cooperative formally twinned under the auspices of the World Organisation for Animal Health (WOAH) to enhance wildlife health capacity in Thailand and the Southeast Asia region. We used a system-wide approach to holistically and interdependently enhance capacity. The project commenced with a wildlife health program needs assessment and capacity enhancement focused on strengthening the general wildlife health surveillance network and improving wildlife health information management. Activities included partner surveys, interactive and didactic workshops, and individual personnel training. Topics included development of wildlife health information management systems, analysis of the current surveillance network, development of a theory of change for a strengthened surveillance network, planning workshops to create a wildlife health network, training on wildlife disease outbreak investigation and field sample collection, leading networks, and individual training on bioinformatics and laboratory techniques. Engagement of stakeholders at all levels, continuous communication throughout the project, use of strategic planning tools and pedagogical methods, and use of iterative and adaptive approaches were key factors to the success of this project.
Keywords: capacity enhancement, One Health, wildlife health
The Integration of One Health Assets to Counter Public Health Threats in the Indo-Pacific
Meghan E. Louis1*
1Public Health Command-Pacific.
*Correspondence: Meghan.e.louis.mil@health.mil
Abstract
The Indo-Pacific region faces significant public health threats that are exacerbated by its complex political and geographic landscape. The U.S. Department of Defense maintains partnerships throughout the region to advance a free and open Indo-Pacific. The Army Public Health Command-Pacific (PHC-P) contains forward stationed public health assets on Army, Airforce, Marine, and Navy installations to maintain the health and readiness of personnel. Public Health Command-Pacific consists of human health, environmental, laboratory, and veterinary services that work together to promote One Health. Each service is integrated into the six Veterinary Readiness Activities, or battalions, in South Korea, Guam (covering Australia and New Zealand), Hawaii, Japan, California, and Washington state (covering Alaska). These Veterinary Readiness Activities are primarily focused on public health, animal medicine, and food protection. In total, PHC-P covers 30 veterinary treatment facilities supporting approximately 370 military working dogs, 700 government-owned animals, and 142,000 privately owned animals. A significant part of the PHC-P mission is food safety and defense, covering commercial sanitary audits for 550 suppliers in over 18 countries, tracking each commodity from farm to table. Other public health assets in the Indo-Pacific include the Armed Forces Research Institute of Medical Sciences. This infectious disease laboratory is the forward-based medical research platform for Asia and Oceana. Both PHC-P and the Armed Forces Research Institute of Medical Sciences strengthen alliances and partnerships within the Indo-Pacific through global health engagements, subject matter expert exchanges, and force health protection exercise support. These opportunities provide excellent collaborative platforms for zoonotic disease surveillance, prevention, and control. Wildlife health and prevention of zoonotic spillover events are critical components of the One Health approach. Collaborative efforts with local and international wildlife health organizations enhance the capacity for comprehensive zoonotic disease surveillance. These partnerships facilitate the sharing of critical data, joint research initiatives, and coordinated response strategies, ensuring a robust defense against zoonotic threats. The integration of wildlife health into the Army’s One Health framework underscores the importance of a holistic approach to public health in the region, reinforcing the commitment to maintain a healthy and ready contingent.
Keywords: Army, public health, global health engagements, wildlife, One Health
New Zealand Biosecurity and Wildlife Surveillance
Kelly Buckle1*
1Biosecurity New Zealand, Tiakitanga Pūtaiao Aotearoa.
*Correspondence: kelly.buckle@mpi.govt.nz
Abstract
New Zealand is an archipelago Pacific nation with a special endemic fauna mostly comprising birds, with a couple of bat species and some marine mammals. The New Zealand MPI is the country’s competent veterinary authority and is responsible for overseeing biosecurity, including surveillance and investigation for exotic diseases of domestic and wild animals. This function occurs under the legislative powers of the Biosecurity Act. When exotic disease is suspected, notifiers are required to contact MPI and are facilitated to do so through a free hotline. Animal disease notifications are assessed on a 24/7 basis by trained veterinary investigators. Notifications are recorded in the national database and either stood down or investigated. If a notification is investigated, we will attempt to investigate the cause of disease to the point of diagnosis, which means ruling out the exotic cause and then doing more testing until a diagnosis is reached. Wildlife notifications are important for multiple reasons including allowing support of One Health, conservation and biodiversity, disease surveillance to facilitate trade, and for social and cultural values. At the end of this presentation, two wild bird mortality investigations were discussed—one that took place in Miranda Pukorokoro (Firth of Thames) involving multiple species, and one in Auckland involving black-billed gulls (tarāpuka). In both investigations, exotic diseases including HPAI was excluded and endemic causes were determined to be the cause of mortality.
Keywords: New Zealand, disease surveillance, animal disease, biosecurity, wildlife
Wildlife Health Risk Management—The Importance of Wildlife Health for One Health Outcomes
Erin M. Davis1*
1Wildlife Health Australia.
*Correspondence: edavis@wildlifehealthaustralia.com.au
Abstract
Effective wildlife health risk management requires a thorough understanding of the drivers of disease risk and an appreciation of the place of wildlife health in One Health; however, the links between biodiversity and health are complex, with biodiversity contributing to health in several ways, including provision of ecosystem services, wellbeing, and food security. Declining biodiversity due to human-induced environmental degradation alters the interactions between host, disease agent, and the environment, increasing the risk of disease spillover. This is further facilitated by global movement of people, animals, and goods; the increasing impacts of climate change; and the ability for disease agents to adapt, resulting in severe impacts on global health, conservation, and socioeconomics when disease outbreaks occur. This highlights the importance of disease risk management strategies that consider the system, integrating wildlife health into One Health approaches throughout; incorporating thoughtful, multisectoral collaboration; and coordinating between animal, human and environmental sectors. Effective and feasible risk management strategies have a greater chance of successful outcomes when risk assessment and management plans are adaptable, incorporate effective risk communication, and use an all-hazards approach and with clear governance, timelines, and clear indicators of success outlined.
There are many examples demonstrating the importance of integrating wildlife health into One Health outcomes. Effective risk communication strategies have led to successful outcomes for wildlife disease risk analysis projects in Australia (Vitali and others, 2023). Wildlife risk communication at a community level is more successful when stakeholders begin well, go local, involve local leaders and groups, tailor interventions to the local culture and language, and when communication is ongoing (MacFarlane and Rocha, 2020). Guidelines developed for community messaging also show how facilitation of effective risk communication can ensure conservation messaging works to neutralize dangerous and unwarranted negative associations between wildlife and disease risk (MacFarlane and Rocha, 2020). Research by Plowright and others (2024) demonstrates the link between ecological countermeasures and prevention of pathogen spillover and pandemic prevention using Hendra virus as a model. Cultural considerations also play a critical role in decision making for the management of wildlife and disease risk. Globally, climate change can impact geographic and temporal patterns of disease, in nonuniform, nonpredictable, and unknowable ways, introducing greater layers of complexity in the management of disease risk involving wildlife, ecosystems, and humans, increasing the value of One Health approaches (Buttke and others, 2021).
There are examples where lack of consideration of wildlife health in management decisions in other sectors led to adverse One Health outcomes. Vulture populations have declined dramatically across the world in the last 30 years, predominantly due to human-led interventions, including use of medications in ungulates that are toxic to vultures, and through other poisoning, hunting, and disease outbreak events (Jalihal and others, 2022). These activities didn’t consider the critical role scavengers play in sanitation, resulting in disease outbreaks in humans (including large numbers of rabies deaths), wildlife, and livestock when vultures were removed from ecosystem (Jalihal and others, 2022). Studies have quantified the social and health costs of wildlife decline (Frank and Sudarshan, 2023). Outcomes like this demonstrate the importance of using a One Health, systems approach that integrates wildlife health for greater understanding of the cause and effect of risk management decisions and for effective disease risk management into the future.
Keywords: disease risk management, One Health, wildlife, risk analysis, risk communication.
Introduction to Korea's Wild Bird Avian Influenza Policy
Suwoong Lee1*
1National Institute of Wildlife Disease Control and prevention.
*Correspondence: hoffman@korea.kr
Abstract
The disease research team of the National Institute of Wildlife Disease Control and Prevention (NIWDC) is mainly responsible for monitoring and response work for wild animal avian influenza (AI), and it also conducts monitoring work for different wild animal diseases such as severe fever with thrombocytopenia syndrome, wild bird diseases (Newcastle Disease virus, West Nile virus, Japanese encephalitis virus, etc.), and wild mammal diseases (foot and mouth disease virus, Marek’s disease virus rabies, coronaviruses, etc.). Among these, the main current disease, wild bird AI, causes enormous social and economic damage every winter in Korea. To systematically respond to such outbreaks, the Avian Influenza Standard Operating Procedure was established, and each organization is taking differentiated measures according to the crisis stage.
During the winter season (September to March), the crisis stage is largely divided into “attention,” “caution,” and “warning” according to the occurrence of HPAI. When HPAI is detected in neighboring countries, the crisis level is the “attention” level, and from October to February of the following year, when winter migratory birds are introduced, it is the “caution” level. In addition, when HPAI is detected in wild birds, it is raised to the “warning” level and relevant measures are taken. In this case, the work of each organization is divided, and a systematic response is taken. The wild bird AI response organizations of the Ministry of Environment are composed of the regional and local basin environmental offices, the National Institute of Biological Resources, the NIWDC, and the Ministry of Environment (headquarters). Among them, the National Institute of Biological Resources investigates the distribution, migration route, and habitat density of winter migratory birds entering the country during the winter, and each regional and local basin environmental office conducts tasks such as monitoring and controlling access to wild bird habitats and feces within the jurisdiction.
The NIWDC conducts AI diagnosis and tests, quickly delivers the results of analyses on the domestic inflow routes and transmission patterns of detected highly pathogenic avian influenza viruses (HPAIV) to relevant organizations. Ultimately, the Ministry of Environment is in charge of comprehensive management and systematic response in wild bird AI. In addition, the NIWDC is discussing and improving relevant policies with related organizations through events such as AI policy forums and AI seminars and also is providing education on AI policies and response measures to local government officials, thereby establishing an organic response system.
Keywords: avian influenza, standard operating procedure, policy
Surveillance of Highly Pathogenic Avian Influenza in Wild Mammals in South Korea
Hyesung Jeong,1* Hyunjun Cho,1 Young-Jae Si,1 Kyunglee Lee,2 and Suwoong Lee1
1National Institute of Wildlife Disease Control and Prevention.
2Cetacean Research Institute of National Institute of Fisheries Science.
*Correspondence: halley@korea.kr
Abstract
Cases of HPAI in wild mammals have been increasing globally. Recently, infections have been confirmed in various wild mammals and livestock across Europe and the Americas, including red foxes, sea lions, American minks, seals, and raccoons, etc. These cases increase the risk of interspecies transmission and human infection, with a high mortality rate of 52 percent posing a significant threat to public health; therefore, monitoring AI in wild mammals is crucial to prevent its spread to livestock and humans and to ensure the health of wildlife. In South Korea, confirmed cases of H5N1 in cats at animal shelters in Yongsan, and Gwanak, Seoul, occurred in July and August 2023. Additionally, detections of HPAI in raptors (eagles, goshawks, etc.) that feed on wild birds are increasing. This study reports the results of surveillance activities conducted to understand the status of HPAI infection in wild mammals in South Korea. The NIWDC of the Ministry of Environment conducted genetic diagnosis and egg inoculation tests on a total of 225 samples (191 terrestrial mammals, 34 marine mammals) collected from April 2023 to March 2024. These samples were obtained through cooperation with wildlife rescue centers, the Cetacean Research Institute of the National Institute of Fisheries Science, and public reports. The terrestrial mammal samples included 132 raccoons, 13 red foxes, 11 otters, 10 weasels, 8 badgers, 2 leopard cats, 2 martens, and 13 other species. The marine mammal samples consisted of 12 finless porpoises, 7 common dolphins, 5 bottle-nosed dolphins, 4 spotted seals, 3 false killer whales, 1 harbor porpoise, 1 Risso dolphin, and 1 sei whale. The results showed that HPAI was not detected in any of the wild mammal samples collected in South Korea; however, given the increasing number of infections in wild mammals overseas, continuous monitoring and surveillance are necessary. It is particularly important to promote public reporting of wild mammal carcasses and to continue the proactive surveillance of HPAI infections to mitigate the risk of transmission. In the event of HPAI confirmation, relevant data will be shared with the Ministry of Environment, the Ministry of Agriculture, Food, and Rural Affairs, the Korea Disease Control and Prevention Agency, and other related agencies for wildlife protection and prevention of diseases in livestock and humans.
Keywords: HPAI, wild mammals, surveillance
Genetic Characterization of Highly Pathogenic Avian Influenza H5N1 and H5N6 Viruses Isolated from Wild Birds in South Korea During the 2023–2024 Winter Season
Young-Jae Si,1 Hyunjun Cho,1 Hyesung Jeong,1* and Suwoong Lee1
1National Institute of Wildlife Disease Control and Prevention.
*Correspondence: mint87@korea.kr
Abstract
Since the initial detection of H5N1 subtype HPAIV in a goose in 1996 in Guangdong, China (Gs/GD), its descendants have spread worldwide, infecting a range of domestic and wild bird species and sporadically spilling over into mammals including humans. The NIWDC in South Korea, has been annually investigating wild bird habitats and major streams in South Korea during winter season as part of AI surveillance. Under the national active surveillance program, fecal specimens and oropharyngeal or cloacal swabs have been collected from wild birds. Carcasses discovered through passive surveillance also have been examined to determine the cause of death in wild birds. During September 2023–March 2024, clade 2.3.4.4b HPAI H5N1 and H5N6 viruses were detected and caused outbreaks in South Korea, including 19 cases in wild birds. To understand the origin of HPAIV, we rapidly conducted complete genome sequencing by next-generation sequencing. Here we performed the phylogenetic analysis of two representative HPAI H5 viruses: A/Eurasian Wigeon/Korea/23WS022-22/2023 [WS022-22/2023(H5N1)] and A/Whooper Swan/Korea/23WC075/2023 [WC075/2023(H5N6)]. Phylogenetic analysis revealed that all 8 viral gene segments of the WS022-22/2023(H5N1) virus shared >99.80 percent nucleotide sequence identity with clade 2.3.4.4b H5N1 viruses identified in Japan during 2023. Maximum likelihood phylogenetic tree showed that all 8 gene segments clustered together with the sequences of HPAI H5N1 viruses identified from wild bird. Meanwhile the HA gene of WC075/2023(H5N6) virus clustered with the H5N1 clade 2.3.4.4b HPAIV that circulated during 2022–23 winter season in South Korea. The NA gene of WC075/2023(H5N6) is closely related to the H5N6 HPAIV from China, which had been isolated from poultry and humans since 2018, but other internal genes were genetically distinct from the H5N6 HPAIV from China. The PB1 and M genes also clustered with 2022–2023 winter season H5N1 HPAIV form Korea. The PB2, PA, NP, and NS genes related to Eurasian low pathogenic avian influenza virus (LPAIV). Estimated time to most recent common ancestor suggests that the novel reassortant H5N6 viruses emerged around August–October 2023 and were subsequently introduced into Japan and Korea. Taken together, wild bird habitats act as hotspots for genetic exchanges of HPAI in wild bird species and contributing to HPAI H5 virus evolution; therefore, our findings highlight that continuous surveillance of HPAI in wild birds is imperative for monitoring the evolution and spread of HPAI via wild birds.
Keywords: HPAI, wild bird, surveillance, phylogenetic analysis, 2023–2024 winter season
Detection of Avian Influenza Virus in Mandarin Duck Since 2020 in South Korea
Kwang-Nyeong Lee,1* Se-Hee An,1 Gyeong-Beom Heo,1 Eun-Kyung Lee,1 and Youn-Jeong Lee1
1Animal and Plant Quarantine Agency.
*Correspondence: leekwn@korea.kr
Abstract
The mandarin duck, Aix galericulata, has become one of the most important species for early detection of HPAIV introduced into South Korea during winter season since 2020. For an AI active surveillance program, more than one thousand live wild birds, resident or migratory, are captured, sampled and tested annually in the Ministry of Agriculture, Food, and Rural Affairs, Republic of Korea. From January 2020 to May 2024, mandarin duck was captured forty times, and, for a single net casting, as many as 30 birds could be captured with 10 on average. Out of the forty capturing events for mandarin duck, we could detect and isolate HPAIV nine times, and, importantly, most of the cases were at the early stage of the four successive HPAI epidemics restricted to every winter season from 2020 to 2024. For the 2023–24 winter season, two subtypes of HPAIV of clade 2.3.4.4b, H5N1 and H5N6, appeared in South Korea from poultry as well as wild birds. On December 4, 2023, we could detect H5N6 HPAIV in two of the 29 mandarin ducks captured in the same net; this was the first detection of H5N6 in wild bird during the 2023–24 winter season following the first report of H5N6 in poultry farm on December 3, 2023. The high susceptibility and detectability of HPAIV in mandarin ducks are believed to be related to their habitat, behavioral characteristics, and interaction with other species, and it is a critical component of the AI surveillance program planning for early detection of AI virus. While LPAIV of H5N3 continued to be detected from early September to December 2023 in wild bird fecal samples and in a broiler duck farm, the H5N3 LPAIV was also detected twice from the captured mandarin ducks in October 2023. It is still not clear what roles the resident or migratory mandarin ducks have for making themselves infected with HPAIV earlier or more vulnerable to HPAI infection.
Keywords: highly pathogenic avian influenza, surveillance, mandarin duck, wild bird, H5N6
Risk-Based Targeted Surveillance for Highly Pathogenic Avian Influenza in Wild Waterfowl in The United States
Ryan S. Miller,1* Krista Dilione,1 Jourdan Ringenberg,1 Sarah N. Bevins,1 and Julie Lenoch1
1U.S. Department of Agriculture.
*Correspondence: ryan.s.miller@usda.gov
Abstract
Designing and implementing disease surveillance systems in wildlife at the national scale is challenging, especially when a system needs to achieve a specific level of detection, achieve early detection, or provide inference related to disease risk in the wildlife population. In the United States a national early detection system for highly pathogenic and other AI viruses of significance in wild migratory birds has been established. To support surveillance efforts in wild waterfowl, an adaptive risk-based approach has been developed that provides surveillance targeting guidance by watershed (drainage basin) and season. To achieve this, we developed a hierarchical Bayesian occupancy model to predict the probability that waterfowl at a site within a watershed are infected with influenza A. The occupancy modeling framework allows integration of spatial and temporal occurrence of influenza A (for example, clustering) and naturally encompasses known seasonal differences in influenza A prevalence in waterfowl and important biological periods for waterfowl migration, both linked to influenza dynamics. The Bayesian model is used annually to identify regions of greatest importance for surveillance. Additionally, we have developed an approach to estimate seasonal watershed-scale waterfowl abundance to allow sample sizes required to achieve detection at the 1 percent design prevalence to be determined.
Highly Pathogenic Avian Influenza Viruses Affecting Alaska Wildlife Exhibit Evidence of Interspecies Transmission and Globally Diverse Recent Common Ancestry
Christina A. Ahlstrom,1* Mia Kim Torchetti,2 Julianna Lenoch,2 Kimberlee Beckmen,3 Megan Boldenow,4 Evan J. Buck,1 Bryan Daniels,4 Krista Dilione,2 Robert Gerlach,5 Kristina Lantz,2 Angela Matz,4 Rebecca L. Poulson,6 Laura C. Scott,1 Gay Sheffield,7 David Sinnett,2 David E. Stallknecht,6 Raphaela Stimmelmayr,7,8 Eric Taylor,4 Alison R. Williams,4 and Andrew M. Ramey1
1U.S. Geological Survey.
2U.S. Department of Agriculture.
3Alaska Department of Fish and Game.
4U.S. Fish and Wildlife Service.
5Alaska Department of Environmental Conservation.
6University of Georgia.
7University of Alaska Fairbanks.
8North Slope Borough.
*Correspondence: cahlstrom@usgs.gov
Abstract
The ongoing panzootic of highly pathogenic H5 clade 2.3.4.4b avian influenza spread to North America in late 2021, with detections of HPAIV in Alaska beginning in April 2022. HPAI viruses have since spread across the state, affecting various species of wild birds, including numerous seabirds as well as domestic poultry and wild mammals. To better understand the dissemination of HPAIV spatiotemporally and among hosts in Alaska and adjacent regions, we compared the genomes of 177 confirmed HPAIV cases detected in Alaska during April through December 2022. Results suggest multiple viral introductions into Alaska during November 2021 through August 2022, as well as dissemination to areas within and outside of the state. We documented evidence for transmission of HPAIV among wild bird species, wild birds and domestic poultry, and wild birds and wild mammals. At least six species of seabirds were affected by diverse HPAIV during 2022, including viruses of four distinct genome constellations sharing recent common ancestry with globally diverse sources. Viral genotypes differed in their spatiotemporal spread, likely effected by timing of introductions relative to population immunity. Continued monitoring for and genomic characterization of HPAIV in Alaska will be important to understand the evolution and dispersal of these economically costly and ecologically relevant pathogens.
Keywords: avian influenza, genomic epidemiology, wildlife
Forecasting Hemispheric-Level Movement of Highly Pathogenic Avian Influenza Resulting from Waterfowl Migration
Wayne E. Thogmartin,1* Ben Goals,1 Andrew M. Ramey,1 Diann J. Prosser,1 and Paul Link2
1U.S. Geological Survey.
2Louisiana Waterfowl Working Group.
*Correspondence: wthogmartin@usgs.gov
Abstract
Understanding migratory waterfowl spatiotemporal distributions is important because, in addition to their economic and cultural value, wild waterfowl can be infectious reservoirs of HPAIV. Waterfowl migration has been implicated in regional and intercontinental HPAIV dispersal, and predictive capabilities for where and when HPAIV may be introduced to susceptible spillover hosts would facilitate biosecurity and mitigation efforts. To develop forecasts for HPAIV dispersal, an improved understanding of how individual birds interact with their environment and move on a landscape scale is required. Using an agent-based modeling approach, we integrated individual-scale energetics, species-specific morphology and behavior, and landscape-scale weather and habitat data in a mechanistic stochastic framework to simulate mallard (Anas platyrhynchos) and northern pintail (Anas acuta) annual migration across the northern hemisphere. Our model recreated biologically realistic migratory patterns using a first principles approach to waterfowl ecology, behavior, and physiology. Conducting a structural sensitivity analysis comparing reduced models to eBird Status and Trends in reference to the full model, we identified density dependence as the main factor influencing spring migration and breeding distributions and wind as the main factor influencing fall migration and overwintering distributions. We show evidence of weather patterns in northeast Asia causing significant intercontinental pintail migration to North America. By linking individual energetics to landscape-scale processes, we identify key drivers of waterfowl migration while developing a predictive model responsive to daily weather patterns. This model paves the way for future waterfowl migration research predicting HPAIV transmission, climate change impacts, and oil spill effects.
Keywords: agent-based modeling, avian physiology, ecological forecasting, mallard, northern pintail, weather
A Simulation Method for Constructing Migratory Host Networks from Band-Recovery and Satellite Telemetry Data
Brooke L. Berger,1* Andy Ramey,2 Ryan Miller,3 Michael Casazza,2 Diann Prosser,2 and Colleen Webb1
1Colorado State University.
2U.S. Geological Survey.
3U.S. Department of Agriculture.
* Correspondence: brooke.berger@colostate.edu
Abstract
Wild waterfowl hosts of influenza A viruses (IAV) can move virus thousands of kilometers during seasonal migrations. To predict where novel subtypes of IAV are likely to move, we need detailed information about host movement. Satellite telemetry and global positioning system trackers provide detailed movement data, but transmitters are expensive and can be difficult to recover, limiting their widespread use. In the United States, bird banding data are far more common due to the low cost of implementation but provide a coarse picture of movement since observations are limited to the initial capture and recovery locations, often with months or years occurring between observations. Our goal was to develop a method that can reconstruct the probable migratory path hosts take between marking and recovery using information gleaned from telemetry studies of fewer individuals. We used satellite telemetry data from 330 northern pintail ducks (Anas acuta) and a previously developed functional movement model to calculate population-level posterior distributions of derived quantities that describe two characteristics of pintail movement: speed and turning angle. Starting at the banding location, we used these quantities and approximate Bayesian computation to simulate thousands of migratory paths for pintails in the USGS North American Bird Banding Dataset, discarding paths that did not end at the known recovery location. This allowed us to probabilistically reconstruct host migration locations while also calculating estimates of uncertainty. We validated our method at the population scale using simulations from telemetered pintail withheld from the derived quantities calculation to generate a spatial migration network, which was compared with a network generated from the real location data. Simulations accurately reconstructed migration paths at the large spatial scale of our analysis and had similar first- and second-order network properties to the observed data network. Our method allows researchers to gain more information about host movement from mark-recapture and mark-recovery data through the integration of telemetry data to better characterize overall movement characteristics.
Keywords: host migration, approximate Bayesian computation, movement simulation, influenza A virus, satellite telemetry, mark-recovery.
Predictors of Influenza A Virus Large-Scale Spatial Transmission and Probable Routes of Viral Movement
Brooke L. Berger,1* Andy Ramey,2 Ryan Miller,3 Nichola Hill,4 Diann Prosser,2 and Colleen T Webb1
1Colorado State University.
2U.S. Geological Survey.
3U.S. Department of Agriculture.
4University of Massachusetts.
*Correspondence: brooke.berger@colostate.edu
Abstract
Influenza A viruses infect highly mobile reservoir hosts allowing viruses to move long distances and exposing them to a variety of habitats and hosts. Understanding how these factors influence the spatial transmission of IAV, and finding the most probable routes of movement, will help us target important areas for surveillance. We developed a network model of spatial transmission from H5 and H7 IAV isolated in 13 different species of wild waterfowl in the contiguous United States. We used the SeqTrack algorithm to construct eight different networks, one for each gene segment, linking viral isolates by their genetic relatedness. These were combined into a single weighted spatial transmission network. We evaluated the impact of temperature, prevalence, host movement, sample delay, and sample effort on spatial transmission between large-scale watersheds and estimated the most probable routes of transmission with a zero-inflated Bayesian binomial network model. Host movement patterns were an important predictor of IAV spatial transmission. Node-level covariates that impact viral detection and environmental persistence did not significantly impact viral movement. The most probable routes of spatial transmission included important breeding habitat for migratory waterfowl and revealed an elevated probability of transmission in the western half of the United States. We present these results here as proof of concept. In August, we will present results from work currently in progress. We apply the same method to low pathogenic IAV isolated in Northern Pintail ducks (Anas acuta) sampled in East Asia and North America. We will examine the impact of the same covariates on spatial transmission among Pintail and highlight possible opportunities for IAV movement between these two continents.
Keywords: spatial transmission, avian influenza, network model, northern pintail ducks
Establishing a Noninvasive Method for Evaluating Susceptibility of Endangered Bird Species to Highly Pathogenic Avian Influenza Virus Using Cultured Cells
Manabu Onuma1
1National Institute for Environmental Studies.
Abstract
We performed infection experiments using cultured cells from 11 bird species to observe responses to highly pathogenic avian influenza viruses. The species included Rock Pigeon, Chicken, Mountain Hawk-Eagle, Northern Goshawk, Peregrine Falcon, Golden Eagle, White-naped Crane, Hooded Crane, Okinawa Rail, Oriental Stork, and Red-crowned Crane. We selected three HPAIV strains—A/chicken/Yamaguchi/7/2004(H5N1); A/black swan Akita/1/2016(H5N6); and A/chicken/Kumamoto/1-7/2014(H5N8)—based on NA subtype variation and reported pathogenicity in chickens. The cells were exposed to the virus for 1 hour, washed with PBS, and then cultured in serum-supplemented KAV-1 medium. Cells were harvested at 6- and 12-hours post-infection, and RNA was extracted to synthesize cDNA. Using primers designed from Mx gene sequences cloned based on the genomic information of the target birds, we measured the temporal expression of the Mx gene.
Our results documented that in the Rock Pigeon, known for its resistance, Mx gene expression increased more than twentyfold over time. In contrast, in chickens, known for high mortality rates, Mx gene expression either decreased or increased only as much as fourfold. Similar Mx gene expression patterns were observed in four raptor species (Mountain Hawk-Eagle, Golden Eagle, Northern Goshawk, and Peregrine Falcon) and the Okinawa Rail compared to chickens. In previous in vivo experiments, raptors showed high mortality or severe neurological symptoms upon HPAIV infection. These findings suggest that the innate immune response, including Mx gene expression, is suppressed in raptor cells when exposed to HPAIV.
Our study highlights the potential to predict organism-level responses to HPAIV based on cellular-level responses. Rock Pigeons, with their large Mx gene expression, show a robust response to HPAIV, aligning with their lower mortality rates. Conversely, the lower and diminishing Mx gene expression in chickens correlates with their higher mortality rates. Raptors and Okinawa Rails exhibit similar Mx gene expression patterns to chickens, indicating a potential vulnerability to HPAIV, as supported by their high mortality and severe symptoms observed in vivo.
The correlation between cellular-level responses and in vivo outcomes indicates the potential to predict HPAIV responses at the organism level from cellular responses. This study suggests that Mx gene expression can be a valuable marker for assessing the susceptibility of different bird species to HPAIV. Further comprehensive gene expression analyses are necessary to elucidate the mechanisms underlying these species-specific differences and to develop effective strategies for managing and mitigating the impact of HPAIV on avian populations. This research contributes to our understanding of AI virus pathology and informs conservation efforts for at-risk species.
Keywords: cultured cell, HPAIV, Mx gene
Strategic Responses to Control and Prevent the Spread of African Swine Fever Virus—Efficient Capture and Surveillance of Wild Boars
Eunji Kang1*
1National Institute of Wildlife Disease Control and Prevention.
*Correspondence: silverg2316@korea.kr
Abstract
Since the first outbreak of African swine fever (ASF) in South Korea in 2019, more than 4,000 ASF-positive wild boars have been confirmed in the past 5 years. Additionally, each year its influence has expanded, occurring in areas equivalent to 28 percent of the national territory as of 2024. ASF primarily spreads through contact among wild boars; therefore, the high density of wild boar population may increase the possibility of accelerating the spread of the ASF virus by direct contact between individuals or groups. So most countries adopt methods such as reducing wild boar populations, removing carcasses, and blocking movement between regions to prevent further spread. In Korea, the population density of wild boars can easily increase because more than 70 percent of the land is mountainous, providing favorable habitat, and there are no predators such as tigers or leopards that prey on wild boars; furthermore, because of the rugged terrain, there are limitations to the human searches needed to thoroughly inspect carcasses for contaminant removal. As ASF outbreaks continue persistently, the NIWDC has developed efficient methods that consider these geographical features. Firstly, we utilize drones equipped with thermal cameras to survey the habitats of wild boars and provide real-time location information about observed wild boars to hunters engaged in nocturnal hunting activities. Secondly, we train detection dogs capable of searching for wild boar carcasses in inaccessible tough terrain where humans cannot enter. This allows us to precisely and swiftly remove contaminants. South Korea has implemented a strategic response approach, particularly focusing on the efficient capture and search of wild boars. And by using this strategy, we will contribute to prevent the spread of ASF though continuous surveillance and prompt response
Keywords: African swine fever virus, wild boar, capture and search
The Efficacy of the African Swine Fever Vaccine Candidate ASFV-G-△I177L/△LVR for Korean Field Virus
Yeonji Kim,1 Won-jun Kim,1 Yongwoo Shin,1 and Weonwha Jheong1*
1National Institute of Wildlife Disease Control and Prevention.
*Correspondence: yeonchi16@korea.kr
Abstract
African swine fever is a highly contagious hemorrhagic disease in domestic pigs and wild boars that results in up to 100 percent case fatality rate. Since the first outbreak in October 2019, the virus has spread out from Gyeonggi-do and Gangwon-do to Chungcheongbuk-do and Gyeongsangbuk-do in South Korea; however, there is no approved vaccine to prevent its spread.
From 2019 to 2020, the U.S. Department of Agriculture (USDA) announced two kinds of vaccine candidates. One is the ASFV-G-△I177L and the other is the ASFV-G-△I177L/△LVR. ASFV-G-△I177L is only replicated in primary swine macrophages cells, making large-scale production impossible; however, ASFV-G-△I177L/△LVR derived from ASFV-G-△I177L replicated effectively and stably in Plum Island porcine epithelial cells. The USDA’s results showed both vaccine candidates have same efficacy and safety for ASFV. In Vietnam, the ASFV-G-△I177L has been partially deployed in recent days; however, there have been reports of fatalities following vaccination. Similar outcomes were observed when we tested ASFV-G-△I177L in domestic pigs.
In this study, we tested ASFV-G-△I177L/△LVR in domestic pigs to confirm the efficacy and safety of the vaccine candidate. We administered it intramuscularly and orally to swine and challenged them with the Korean field virus (isolated from Hwacheon in 2020; Genbank accession: OR15927.1). To assess the safety of the vaccine candidate in late-stage sows, we administered it and evaluated the outcomes of offspring delivery, antibody formation, and transmission of maternal antibodies to fetuses through colostrum. Overall, these results suggest that ASFV-G-△I177L/△LVR could be a potential candidate against the current African swine fever virus (ASFV) outbreak; however, more experiments need to be conducted to confirm the safety of the vaccine candidate.
Development and Characterization of High-Efficiency Cell-Adapted Live Attenuated Vaccine Candidate Against African Swine Fever Virus
Sung-Sik Yoo,1 In Pil Mo,2 Weonhwa Jheong,3 and Jong-Soo Lee4*
1ChoongAng Vaccine Laboratories.
2AviNext, Cheongju.
3National Institute of Wildlife Disease Control and Prevention.
4Chungnam National University.
*Correspondence: jongsool@cnu.ac.kr
Abstract
African swine fever is a contagious and lethal hemorrhagic disease that occurs in domestic pigs and wild boars and poses a significantly threat to the global pig industry. Several experimental vaccine candidates derived from naturally attenuated, genetically engineered, or cell culture-adapted Asian swine fever virus (ASFV) have been tested, but no commercial vaccine is accepted globally. Here, we developed a safe and effective cell-adapted live attenuated vaccine candidate (ASFVMEC-01) by serial passaging of a filed isolate in the CA-CAS-01-A cell. The ASFV-MEC-01 strain was obtained at passage level 18 and showed significant attenuation in 4- and 6-week-old pigs. In particular, the ASFV-MEC-01 strain conferred 100 percent protection against lethal parental ASFV challenge in immunized pigs, which correlated with high ASFV-specific humoral and cellular immune responses. Additionally, the ASFV-MEC-01 strain was not detected from blood until 28 days post-inoculation. Global transcriptome analysis showed that the ASFV-MEC-01 strain with a deletion of 12 genes in the left variable region had more innate antiviral properties than parental virus, as exemplified by high mRNA levels of interferon regulatory and inflammatory genes in primary porcine alveolar macrophages. Ectopic expression of most deleted genes enhanced replication of DNA viruses by suppressing the production of interferons (IFNs) and pro-inflammatory cytokines. These results suggest that attenuation of the ASFV-MEC-01 strain may be mediated by the induction of higher type I IFN and NF-κB signaling from the host innate immune system. Collectively, the ASFV-MEC-01 strain could be a safe and effective live attenuated ASFV vaccine candidate with commercial potential.
Keywords: African swine fever, CA-CAS-01-A cell, live attenuated vaccine, ASFV-MEC-01, type I IFN
African Swine Fever Prevention and Preparedness Activities Targeting Feral Swine in the United States
Dana Cole,1* Vienna Brown,1 and Michael Marlow1
1U.S. Department of Agriculture.
*Correspondence: dana.j.cole@usda.gov
Abstract
African swine fever, caused by ASFV, is a hemorrhagic disease that results in high levels of morbidity and mortality among members of the Suidae family. The disease has recently emerged as a significant global threat to the pork industry, in part due to introduction and transmission among wild pig populations in several countries. Many characteristics of ASFV make it particularly well suited for an epizootic event including high virulence, low infectious dose, systemic dissemination throughout the body of the host, and extensive viral shedding. In addition, the ASFV is highly stable in pork and pork products despite exposure to varying temperatures and pH during common methods of meat curing, consequently, in response to the detection of ASFV in Hispaniola in 2021, the United States increased preparedness and prevention planning activities for a potential ASFV entry, including a focus on invasive feral swine.
Because of the unique risk of entry and introduction to free-roaming and feral swine in the U.S. territories—Puerto Rico and the U.S. Virgin Islands—enhanced removal and surveillance activities commenced quickly in hard-hit areas. Over time, risk-based, targeted removal and surveillance of invasive feral swine increased in the continental United States, and now surveillance in feral swine spans the entire invaded range. In addition to the ASF response plan red book, an ASF response playbook for feral swine was released in July 2023. In 2024, the United States continues to enhance international and national partnerships to increase readiness and response coordination and is building additional capacity to trap and remove feral swine if an outbreak of ASFV is detected in invasive feral swine.
Ten Years of Feral Swine (Sus scrofa) Disease Surveillance in Guam
Charlene B. Hopkins,1* D. Buck Jolley,1 and Aaron F. Collins1
1U.S. Department of Agriculture.
*Correspondence: charlene.hopkins@usda.gov
Abstract
Samples from 3,358 feral swine (Sus scrofa) in Guam have been collected and tested for various diseases from June 2014 to May 2024. In 2014, 13 samples from feral swine in Guam were collected by the USDA Animal and Plant Health Inspection Service, Wildlife Services for disease testing. Early samples from feral swine were tested for 12 diseases. By 2017, sample collection for disease surveillance became routine and six diseases were under surveillance. Then by 2019, disease surveillance included three diseases (swine brucellosis, pseudorabies virus, and classic swine fever). Visual inspection of feral swine for signs of foot and mouth disease was added to the disease surveillance program in 2021, and in April 2024 testing for ASF was added. Positive detections have been found for swine brucellosis, pseudorabies virus, influenza A virus, leptospirosis, toxoplasmosis, trichinellosis, and Japanese encephalitis virus in Guam feral swine samples. Classic swine fever, bluetongue virus, porcine epidemic diarrhea virus, porcine reproductive and respiratory syndrome, tuberculosis, and ASF test results have been negative thus far. Signs of foot and mouth disease have not been detected. Prevalence of swine brucellosis and pseudorabies has fluctuated throughout time, and prevalence is generally higher than in the continental United States. Swine brucellosis and pseudorabies virus are detected throughout Guam.
Keywords: feral swine, disease monitoring, swine brucellosis, pseudorabies virus, classic swine fever, foot and mouth disease, African swine fever, influenza A virus, leptospirosis, toxoplasmosis, trichinellosis, Japanese encephalitis virus, disease prevalence
Adaptive Risk-Based Targeted Surveillance for Foreign Animal Diseases in Wild Pigs
Ryan S. Miller,1* Mary J. Woodruff,1 Greg Franckwiak,1 Vienna R. Brown,1 and Dana Cole1
1U.S. Department of Agriculture.
*Correspondence: ryan.s.miller@usda.gov
Abstract
Animal disease surveillance is an important component of the national veterinary infrastructure to protect animal agriculture and facilitates identification of foreign animal disease (FAD) introduction. Once introduced, pathogens shared among domestic animals and wildlife are especially challenging to manage due to the complex ecology of spillover and spillback; thus, early identification of FAD in wildlife, especially wild pigs, is critical to minimize outbreak severity and potential impacts to animal agriculture as well as potential impacts to wildlife and biodiversity. As a result, national surveillance and monitoring programs that include wildlife are becoming increasingly common. Designing surveillance systems in wildlife or, more importantly, at the interface of wildlife and domestic animals is especially challenging because of the frequent lack of ecological and epidemiological data for wildlife species and technical challenges associated with a lack of noninvasive methodologies. To meet the increasing need for targeted FAD surveillance in wild pigs, we developed an adaptive risk-based targeted surveillance approach that accounts for risks in source and recipient host populations. The approach is flexible, accounts for changing disease risks through time, can be scaled from local to national extents, and permits the inclusion of quantitative data or, when information is limited, to expert opinion. We apply this adaptive risk-based surveillance framework to prioritize areas for surveillance in wild pigs in the United States with the objective of early detection of three diseases, classical swine fever, ASF, and foot-and-mouth disease. We discuss our surveillance framework, its application to wild pigs, and discuss the utility of this framework for surveillance in other host species and diseases.
Antimicrobial Resistance and One Health—An Ecological Perspective
Alan B. Franklin1*
1U.S. Department of Agriculture.
*Correspondence: alan.b.franklin@usda.gov
Abstract
Ideally, One Health is a transdisciplinary approach that encompasses human medicine, veterinary medicine, and environmental and ecological sciences; however, One Health research is often conducted solely within the human and veterinary medical professions and often does not include the environmental or ecological component. This is especially true when attempting to understand antimicrobial resistance (AMR) where research is often siloed into either human medicine or veterinary medicine and rarely encompasses both disciplines. Although wildlife is increasingly recognized as potential components in AMR in the environment, cross disciplinary research coupling ecological science with human and veterinary medicine is rare. As an example, we use a research program on the ecological role of wildlife in maintaining and transmitting AMR bacteria at the wildlife-agricultural and wildlife-human interfaces. This research program included field sampling wildlife around two primary sources of AMR (wastewater treatment plants and livestock feedlots), conducting experimental infections of wildlife as hosts of AMR, and tracking movements of wildlife infected with AMR bacteria. We found evidence that wastewater treatment plants and livestock facilities were sources of AMR for wildlife and that different wildlife species can serve as maintenance and bridge hosts for AMR. Wildlife infected with AMR also can move AMR from local scales (for example, among agricultural operations) to continental scales (for example, between North America and Asia). To better integrate our research into a One Health approach, we have enlisted collaborators from the medical, veterinary, and environmental disciplines in a large-scale study designed to elucidate the proximate and ultimate sources of AMR in wildlife and the natural environment and the implications to human and agricultural health.
Keywords: antimicrobial resistance, One Health, wildlife, human health, agricultural health
Pathogen Spillover—From Wildlife Reservoirs to Global Epidemics
Hongxuan He1*
1Chinese Academy of Sciences.
*Correspondence: hehx@ioz.ac.cn
Abstract
Since the outbreak of the coronavirus disease 2019 (COVID-19) pandemic, the spillover of pathogens from animals to humans within the wildlife trade has garnered significant attention. The growing human population and expanding agricultural activities have increased interactions between humans and wildlife reservoirs. These interactions are often accompanied by behaviors that elevate the likelihood of spillover, such as the consumption of wild meat or the enhanced contact between wildlife and domestic animal hosts. For novel pathogens to pose a significant threat to human populations, initial contact between human and animal hosts is essential; moreover, the pathogen must have the capability to infect humans or undergo evolutionary changes that enable human-to-human transmission. Crucially, this interpersonal transmission must extend beyond the initial outbreak location to pose a widespread threat. Changes in global connectivity have introduced a distinct risk for pathogen transmission, facilitating their spread more extensively and rapidly than ever before. In our analysis, we use the influenza virus as a case study to illustrate the complexity of pathogen transmission across species. In an increasingly interconnected world, the risks of new zoonotic diseases emerging from wild animals are shared globally, underscoring the necessity for a comprehensive approach to detect, monitor, prevent, and control infectious diseases. To address these challenges, we provide comprehensive disease control recommendations. The One Health policy is instrumental in investigating the origins and potential sources of new zoonotic diseases, offering crucial information to develop policy strategies to confront the emergence of new infectious diseases.
Keywords: pathogen, spillover, wildlife, influenza, one health
Applying Biomedical Tools to Understand Coral Disease
Thierry M. Work1*
1U.S. Geological Survey.
*Correspondence: Thierry_work@usgs.gov
Abstract
Coral reefs globally are declining because of human-caused insults. Land-based pollution is smothering reefs, and global warming is leading to increased bleaching events where symbiotic algae leave the coral host, turning them white leading to coral death. Now, disease is emerging as a substantial threat to corals. A recent example is stony coral tissue loss disease (SCTLD) that started in Florida (USA) and is now spreading throughout the entire Caribbean. There is concern that SCTLD or something similar could impact corals reefs in the Pacific, yet managers have few tools to respond, in part because infrastructure to respond to and investigate causes of disease in corals is lacking. Here we highlight efforts to develop tools and capacity to respond to and investigate disease in corals using a systematic approach in the Pacific and Caribbean. The strategy hinges on coordinating field and laboratory response. For the field component, biologists are trained to systematically describe lesions in corals, to recognize disease outbreaks, and to properly procure samples for laboratory analysis. The lab component is focused on systematically describing lesions at the gross and microscopic levels to rule in or out potential causative agents of disease. In addition, tools used to investigate disease in other animals are being adapted to corals to understand disease pathogenesis in this enigmatic group of organisms. This approach has led to several breakthroughs including standardized methods to describe lesions, standardized field manuals to investigate disease, and the discovery that viruses may play a role in SCTLD in Florida.
Keywords: coral reefs, epizootiology, disease causation, microscopy
Understanding Drivers of Winter Tick (Dermacentor albipictus) Abundance and Distribution in New England
Tammy L. Wilson,*1,2 Juliana Berube,2 Alexej P. K. Sirén,2 Benjamin Simpson,3 and Kelly Klingler2
1U.S. Geological Survey.
2University of Massachusetts.
3Penobscot Nation.
*Correspondence: tammywilson@umass.edu
Abstract
Moose (Alces alces; Mos in Algonquin [Penobscot] language) are an important symbolic and subsistence animal for native peoples in North America. Over the past several decades, moose populations in New England have been in decline due to winter tick (Dermacentor albipictus) parasitism. Although not known to carry common tick-borne pathogens, winter tick infestations of moose can be severe, and epizootics have been associated high annual calf mortality rates and reductions in annual fecundity. We used winter tick abundance and distribution collected on- and off-host to evaluate the potential drivers of winter tick epizootics throughout the northeastern United States. We found that imperfect detection was common when sampling off-host winter ticks using a modified flagging technique. Detection probability peaked in the middle of October, and tick occupancy and abundance were highest in intermediate understory vegetation height. Parasite loads on moose peaked at mid-latitudes and are associated with warmer and wetter summers before questing. Region-wide deployment of rigorous sampling is important to gain an accurate picture of winter tick population dynamics. By focusing on the ecology of tick distribution and abundance we hope to design effective mitigation strategies that will minimize the risk to moose populations.
Use of Riparian Habitat by Invasive Culex quinquefasciatus and the Fate of Hawaiian Honeycreepers at Hakalau Forest National Wildlife Refuge, Island of Hawai’i
Dennis A. LaPointe,1* Mei N. Iwamoto,2 Matthew C. Mueller,2 Katherine M. McClure,1 Richard J. Camp,1 and Lucas Berio Fortini1
1U.S. Geological Survey.
2University of Hawai’i at Hilo.
*Correspondence: dlapointe@usgs.gov
Abstract
Avian malaria, Plasmodium relictum, is the key limiting factor driving extinctions and population declines of Hawaiian honeycreepers. It is transmitted by the invasive southern house mosquito, Culex quinquefasciatus, which has become established in forest bird habitat. In the past two decades, honeycreeper populations have declined significantly on Kauaʻi and Maui, but they remain relatively stable at Hakalau Forest National Wildlife Refuge on Hawaiʻi Island. Limited availability of suitable larval habitat may be one reason Culex quinquefasciatus are seldom detected at Hakalau and why avian malaria remains at low prevalence in the avian community. Changing patterns in precipitation and stream hydrology, however, could increase riparian larval habitat and provide a stepping-stone invasion route into higher elevation forests like Hakalau. We established seven sites on an elevation gradient (1110–1750 meters) along two streams in Hakalau and one stream in the adjacent Laupāhoehoe Forest Reserve and conducted regular surveys for available larval habitat and larval mosquito presence. We also established climate stations and stream gages at our sites to collect key environmental data. We found available larval habitat in the form of rock holes and perched pools, and Aedes japonicus larvae in all streams and at all elevation sites. We found Culex quinquefasciatus larvae at all elevation sites along the Laupāhoehoe Forest Reserve stream but, despite abundant and persistent available habitat and presence of Aedes japonicus, we found no Culex quinquefasciatus larvae in streams in Hakalau Forest National Wildlife Refuge. Preliminary analysis of climate and hydrological data suggest cooler water temperatures and shorter periods between high water events may have prevented the establishment of Culex quinquefasciatus in the refuge. Our larval mosquito, larval habitat, and environmental data will be used to assess future risk of Culex quinquefasciatus invasion and avian malaria transmission at Hakalau Forest National Wildlife Refuge.
Keywords: Hawaiian honeycreepers, avian malaria, Culex quinquefasciatus, climate change, riparian habitat
Ke Kai Ola “The Healing Sea”—10 Years of Hawaiian Monk Seal Conservation Effort
Sophie Whoriskey,1* Megan McGinnis,1 Cara L. Field,1 Dominic Travis,1 and Michelle Barbieri2
1Ke Kai Ola, The Marine Mammal Center.
2National Oceanic and Atmospheric Administration.
*Correspondence: whoriskeys@tmmc.org
Abstract
The Hawaiian monk seal (Neomonachus schauinslandi) is endemic to the Hawaiian archipelago and the only extant tropical phocid alive today. The Hawaiian monk seal population has grown roughly 2 percent per year during the past decade after decades of monitoring and mitigation to support recovery; however, these seals continue to face numerous threats to their survival across the archipelago with most of the threats related to human activities, including fisheries interactions, intentional killings, entanglements, and climate change-associated habitat loss. Over the last 10 years, The Marine Mammal Center, in collaboration with NOAA Fisheries, has operated a Hawaiʻi-based multidisciplinary program to address these threats through rehabilitation and health monitoring of individual Hawaiian monk seals and through public education and outreach. A total of 50 individual Hawaiian monk seals have been rehabilitated through this program, representing approximately 3 percent of the population, with 92 percent (n=46) of these animals successfully released. The majority of these patients were severely malnourished young seals under 2 years of age, with additional animals being admitted for trauma, accidental hook ingestion, and the disease toxoplasmosis. Long-term survival and reproductive success of released patients is being investigated to better understand the impact of rehabilitation as a conservation tool for this species. In addition to rehabilitation, conservation efforts have targeted increasing public awareness and knowledge of monk seals and addressed specific threats such as entanglement, accidental hooking in fishing gear, and other human interactions that affect this species and their ecosystem. These interactions are particularly important in the main Hawaiian Islands where the roughly 1.5 million residents from diverse communities with great variation in culture, geography, and economic status meet the approximately 9 million visitors each year in sharing the coastal ocean environment that monk seals depend upon to survive. Public outreach efforts have reached thousands of residents and visitors, and school programs have reached hundreds of students annually. Similar to our ongoing investigation of monk seal rehabilitation success and contribution to the population, current program efforts are underway to evaluate measures of success and to identify areas in which to further develop and improve.
Keywords: Hawaiian monk seal, wildlife, health, rehabilitation
Current Status and Response to ASF Outbreaks in Wild Boars in Korea
Weonhwa Jheong1*
1National Institute of Wildlife Disease Control and Prevention.
*Correspondence: purify@korea.kr
Abstract
African swine fever is a disease with a mortality rate of about 100 percent that infects only domestic pigs and wild boars. When ASF spreads to a region or a country, it greatly impacts the wild boar ecosystem and causes significant damage to the pig farming industry, as it is a disease that requires the culling of all pigs on affected farms. The ASF outbreak that started in the country of Georgia in 2007 is still ongoing in Europe and Asia. In Korea, ASF was first detected in a pig farm on September 16, 2019, and in wild boars in early October of the same year. To date, more than 4,000 cases have been reported over a period of 57 months. Korea is responding to the ASF outbreak through measures such as carcass search and removal, strengthening capture efforts to reduce wild boar populations, installing fences to restrict wild boar movement, and monitoring environmental contamination. Unlike in Europe, Korea’s wild boar habitats are mainly in high and rugged mountainous areas, making response efforts more challenging. In the early stages of the wild boar ASF outbreak in 2020 and 2021, Korea focused on intensive monitoring and capture efforts around affected areas. Despite these efforts, the spread continued, leading to the implementation of a nationwide continuous monitoring system in 2022. Additionally, to curb the natural spread of ASF, Korea intensified capture efforts to lower the wild boar density to below 1.0 boar/square kilometer (km2; with a maximum target of 0.7 boar/km2). Since 2023, carcass detection dogs and thermal observation drones have been introduced to efficiently find location and capture wild boars, and measures have been taken to prevent illegal activities such as the unauthorized use of hunting dogs and the movement of wild boar carcasses, as well as to prevent artificial contamination during hunting and sampling activities.
Keywords: wild boar ASF, carcass searching dog, TOD (thermal observation device), artificial contamination, illegal (cheating) act
References Cited
Adisasmito, W.B., Almuhairi, S., Behravesh, C.B., Bilivogui, P., Bukachi, S.A., Casas, N., Cediel Becerra, N., Charron, D.F., Chaudhary, A., Ciacci Zanella, J.R., Cunningham, A.A., Dar, O., Debnath, N., Dungu, B., Farag, E., Gao, G.F., Hayman, D.T.S., Khaitsa, M., Koopmans, M.P.G., Machalaba, C., Mackenzie, J.S., Markotter, W., Mettenleiter, T.C., Morand, S., Smolenskiy, V., and Zhou, L., 2022, One Health—A new definition for a sustainable and healthy future: PLoS Pathogens, v. 18, no. 6, p. e1010537, accessed August, 24, 2024, at https://doi.org/10.1371/journal.ppat.1010537.
Buttke, D., Wild, M., Monello, R., Schuurman, G., Hahn, M., and Jackson, K., 2021, Managing wildlife disease under climate change: EcoHealth, v. 18, no. 4, p. 406–410, accessed August 24, 2024, at https://doi.org/10.1007/s10393-021-01542-y.
Frank, E., and Sudarshan, A., 2023, The social costs of keystone species collapse—Evidence from the decline of vultures in India: Becker Friedman Institute for Economics Working Paper No. 2022-165, 72 p., accessed August 24, 2024, at https://doi.org/10.2139/ssrn.4318579.
Jalihal, S., Rana, S., and Sharma, S., 2022, Systematic mapping on the importance of vultures in the Indian public health discourse: Environmental Sustainability, v. 5, no. 2, p. 135–143, accessed August 24, 2024, at https://doi.org/10.1007/s42398-022-00224-x.
MacFarlane, D., and Rocha, R., 2020, Guidelines for communicating about bats to prevent persecution in the time of COVID-19: Biological Conservation, v. 248, p. 108650, accessed August 8, 2024, at https://doi.org/10.1016/j.biocon.2020.108650.
Pepin, K.M., Carlisle, K., Anderson, D., Baker, M.G., Chipman, R.B., Benschop, J., French, N.P., Greenhalgh, S., McDougall, S., Muellner, P., Murphy, E., O’Neale, D.R.J., Plank, M.J., and Hayman, D.T.S., 2024, Steps towards operationalizing One Health approaches: One Health, v. 18, p. 100740, accessed August 24, 2024, at https://doi.org/10.1016/j.onehlt.2024.100740.
Plowright, R.K., Ahmed, A.N., Coulson, T., Crowther, T.W., Ejotre, I., Faust, C.L., Frick, W.F., Hudson, P.J., Kingston, T., Nameer, P.O., O’Mara, M.T., Peel, A.J., Possingham, H., Razgour, O., Reeder, D.A.M., Ruiz-Aravena, M., Simmons, N.B., Srinivas, P.N., Tabor, G.M., Tanshi, I., Thompson, I.G., Vanak, A.T., Vora, N.M., Willison, C.E., and Keeley, A.T.H., 2024, Ecological countermeasures to prevent pathogen spillover and subsequent pandemics: Nature Communications, v. 15, no. 1, p. 2577, accessed August 24, 2024, at https://doi.org/10.1038/s41467-024-46151-9.
Vitali, S.D., Reiss, A.E., Jakob-Hoff, R.M., Stephenson, T.L., Holz, P.H., and Higgins, D.P., 2023, National Koala Disease Risk Analysis Report, version 1.2: University of Sydney web page, accessed August 24,2024, at https://wildlifehealthaustralia.com.au/Resource-Centre/Biosecurity-Management.
Appendix 1. Asia-Pacific Wildlife Health Workshop 2024
Goals and Workshop Themes
-
• To continue to build the wildlife health community of practice in the Asia-Pacific region and expand the participants to agencies and institutions from other countries in the region.
-
• Exchange scientific knowledge among the participants to share best practices, create scientific networks, and build capacity in wildlife health science for the Asia-Pacific region.
The themes for this workshop are:
Abbreviations
AI
avian influenza
AMR
antimicrobial resistance
ASEAN
Association of Southeast Asian Nations
ASF
African swine fever
ASFV
African swine fever virus
COVID-19
coronavirus disease 2019
FAD
foreign animal disease
FP
fibropapillomatosis
HPAI
highly pathogenic avian influenza
HPAIV
highly pathogenic avian influenza viruses
IAV
influenza A viruses
IFN
interferons
LPAIV
low pathogenic avian influenza virus
MAL
Ministry of Agriculture and Livestock
MPI
Ministry of Primary Industry
NIWDC
National Institute of Wildlife Disease Control and Prevention
NOAA
National Oceanic and Atmospheric Administration
Para-vet
Para-veterinarian
PHC-P
The Army Public Health Command-Pacific
SCTLD
stony coral tissue loss disease
SPC
Pacific Community
TEIDs
transboundary emerging infectious diseases
USDA
U.S. Department of Agriculture
USGS
U.S. Geological Survey
WDRA
wildlife disease risk analysis
WHA
Wildlife Health Australia
WOAH
World Organisation for Animal Health
For more information about this publication, contact:
Director, USGS Midcontinent Region
1992 Folwell Ave.
St. Paul, MN 55108
For additional information, visit: https://www.usgs.gov/regions/midcontinent
Publishing support provided by the
Rolla Publishing Service Center
Disclaimers
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Although this information product, for the most part, is in the public domain, it also may contain copyrighted materials as noted in the text. Permission to reproduce copyrighted items must be secured from the copyright owner.
Suggested Citation
Sleeman, J.M., comp., 2025, Proceedings of the 2024 Asia-Pacific Wildlife Health Workshop—Collaborating against shared threats: U.S. Geological Survey Open-File Report 2024-1081, 23 p., https://doi.org/10.3133/ofr20241081.
ISSN: 2331-1258 (online)
| Publication type | Report |
|---|---|
| Publication Subtype | USGS Numbered Series |
| Title | Proceedings of the 2024 Asia-Pacific Wildlife Health Workshop—Collaborating against shared threats |
| Series title | Open-File Report |
| Series number | 2024-1081 |
| DOI | 10.3133/ofr20241081 |
| Publication Date | January 30, 2025 |
| Year Published | 2025 |
| Language | English |
| Publisher | U.S. Geological Survey |
| Publisher location | Reston, VA |
| Contributing office(s) | Midcontinent Regional Director's Office |
| Description | vii, 23 p. |
| Online Only (Y/N) | Y |
| Additional Online Files (Y/N) | N |